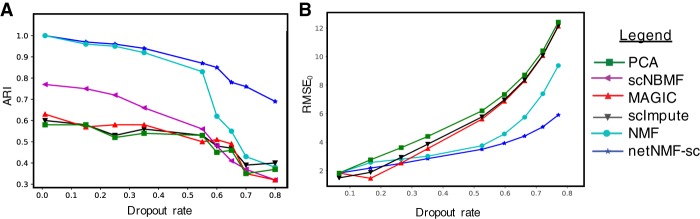
Figure 2.
Comparison of netNMF-sc and other methods on a simulated scRNA-seq data set containing 1000 cells and 5000 genes, with dropout simulated using a multinomial dropout model. (A) Adjusted Rand Index (ARI) between the true and inferred cell clusters obtained as a function of dropout rate. (B) RMSE at dropped-out entries (RMSE0) between true and imputed transcript counts.