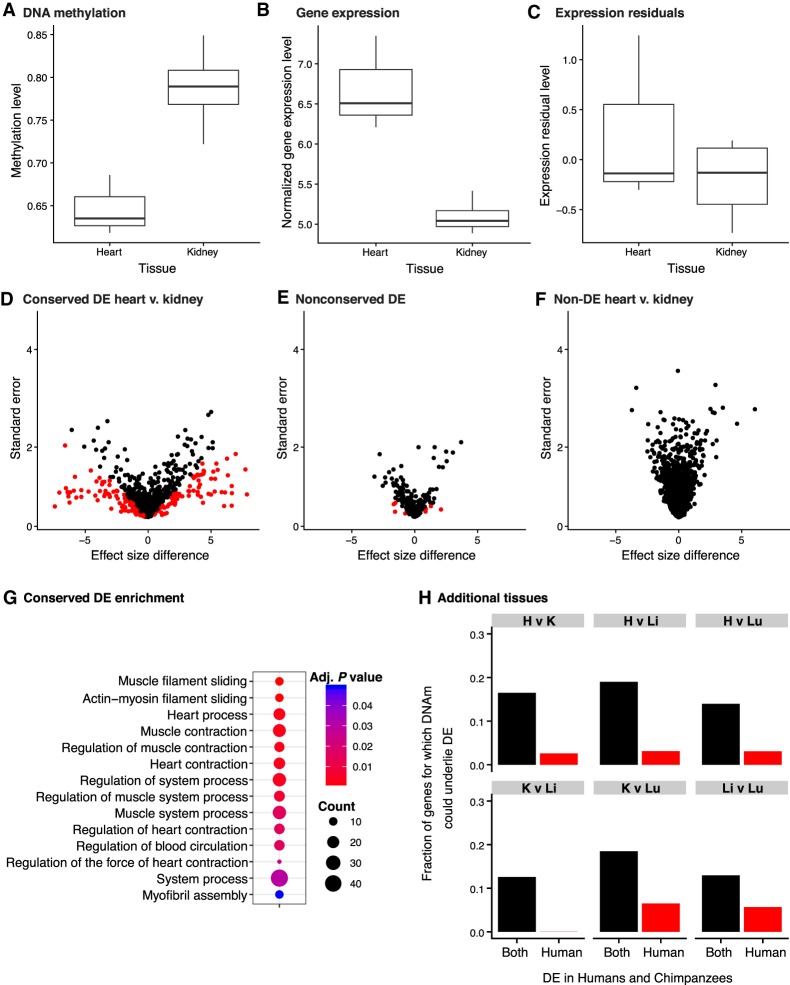
Figure 4.
Inter-tissue DNA methylation and gene expression levels (FDR = 0.05 and FSR = 0.05). (A–C) A representative example of the PRKACA gene in which the variation of methylation levels (A) may explain the differences in gene expression levels (B) between human heart and kidney. (C) The residuals of normalized gene expression levels after regressing out methylation levels. (D–G) Next, we compared the tissue effect sizes before and after controlling for DNA methylation levels in inter-tissue DE and non-DE genes, separately. Genes in red are significant at S-value <0.05. Effect size differences in conserved DE genes in human heart relative to human kidney (D), nonconserved DE genes in human heart relative to human kidney (E), and non-DE genes in human heart and human kidney (F). (G) The conserved DE genes are enriched for heart-related function. (H) Variation in DNA methylation is more likely to explain variation in conserved DE genes than nonconserved DE genes (DE in the human tissues listed, but not in the same tissues in chimpanzees).