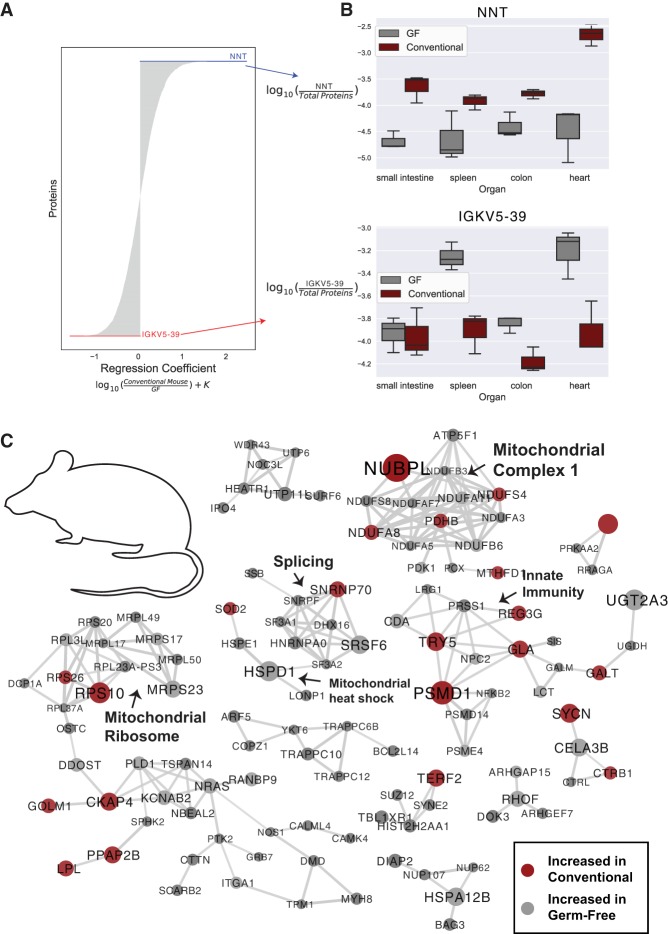
Figure 4.
Organism-level protein networks modulated by the microbiome. A multinomial regression controlling for organ and microbial colonization state of the mice was used to assess proteins associated with colonization status. (A) Proteins ranked by regression coefficient; proteins with coefficients of the greatest magnitude are most associated with colonization status. Proteins with positive coefficients are more abundant in conventional mice, while proteins with negative coefficients are more abundant in GF mice. (B) Log abundance of NNT or IGKV5-39 over the entire proteome in each organ. (C) Protein–protein interaction networks from the top-ranked proteins from the multinomial regression associated with both conventional and GF status when controlling for organ and mouse. The top 150 proteins associated with both GF and conventional status were analyzed (300 proteins total), and proteins with high confidence interactions (0.8) are shown. Nodes are sized by the absolute value of the regression coefficient and colored by association with GF (gray) or conventional (red) status. Putative functional groupings are indicated.