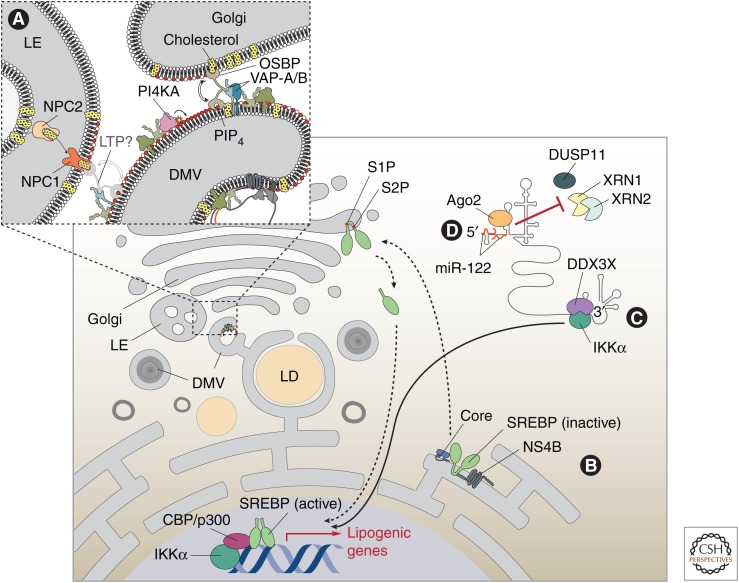
Figure 3.
Exploitation of lipid pathways and miR-122 by hepatitis C virus (HCV). (A) HCV alters the lipid composition of rearranged membranes. NS5A and NS5B recruit and activate phosphatidylinositol 4-kinase-α (PI4KA) to produce a local accumulation of phosphatidylinositol 4-phosphate (PI4P). This may determine the directionality of cholesterol transfer by lipid transfer proteins (LTPs), such as oxysterol-binding protein (OSBP), which is recruited by NS5A via VAP-A/B and releases cholesterol in exchange for PIP4 at these membrane contact sites. VAP proteins might serve as anchors for additional host proteins promoting the formation of endoplasmic reticulum (ER)–late endosome (LE) membrane contacts. Here NPC1, possibly in coordination with NPC2, mediates the export of unesterified cholesterol that might be accepted by lipid transfer proteins recruited by HCV (indicated with a question mark). (B,C) HCV infection activates the transcription of lipogenic genes by two distinct pathways. (B) The inactive SREBP precursor traffics from the ER to the Golgi on HCV infection or expression of core or NS4B. There, the transcriptionally active amino-terminal segment is released after two-step proteolytic processing by the site 1 protease (S1P) and S2P. Upon dimerization, the active SREBP enters the nucleus and activates the transcription of lipogenic genes. (C) The HCV 3′UTR interacts with DEAD box polypeptide 3X-linked (DDX3X). This RNA-binding protein activates IKK-α, which stimulates CBP-p300 to promote SREBP-mediated transcription. (D) miR-122, in association with Agonaute-2 (Ago2), binds to the HCV 5′UTR and protects the viral genome from 5′ triphosphate removal and nucleolytic degradation by 5′-3′ exoribonucleases 1 (XRN1) and XRN2.