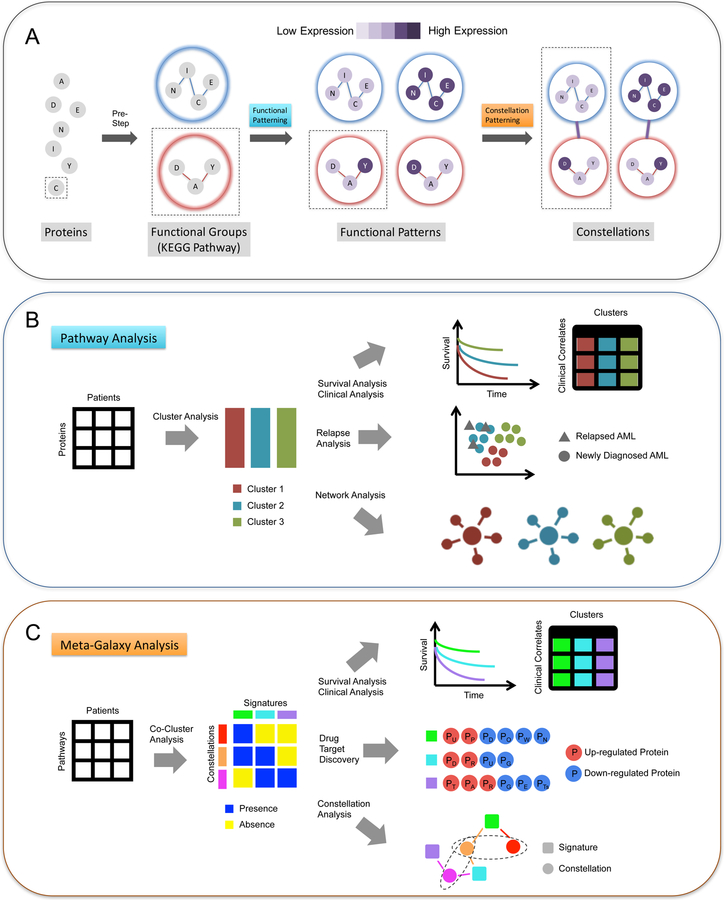
Figure 1. MetaGalaxy analysis workflow.
The schematic of (A) the overall workflow of MetaGalaxy analysis: proteins are organized into functional groups from known biology; analysis identifies patterns of protein expression for the functional groups (Functional Patterning); and finally, constellations, or patterns across functional groups are identified (Constellation Patterning). (B) steps in Functional Patterning include identifying the optimal groups for functional patterns via Progeny Clustering of the protein expression levels for the 209 AML patients; survival, relapse and other clinical covariate analyses; and signaling network analysis; and (C) steps in Constellation Patterning include co-clustering to identify which patients fall into each protein functional pathway (signatures) and how protein functional pathways are grouped (constellations); survival and other clinical analyses; drug target discovery for proteins that are significantly up- or down-regulated across patient groups; and decision tree modeling to identify the contribution of each protein constellation to patient outcomes (constellation analysis).