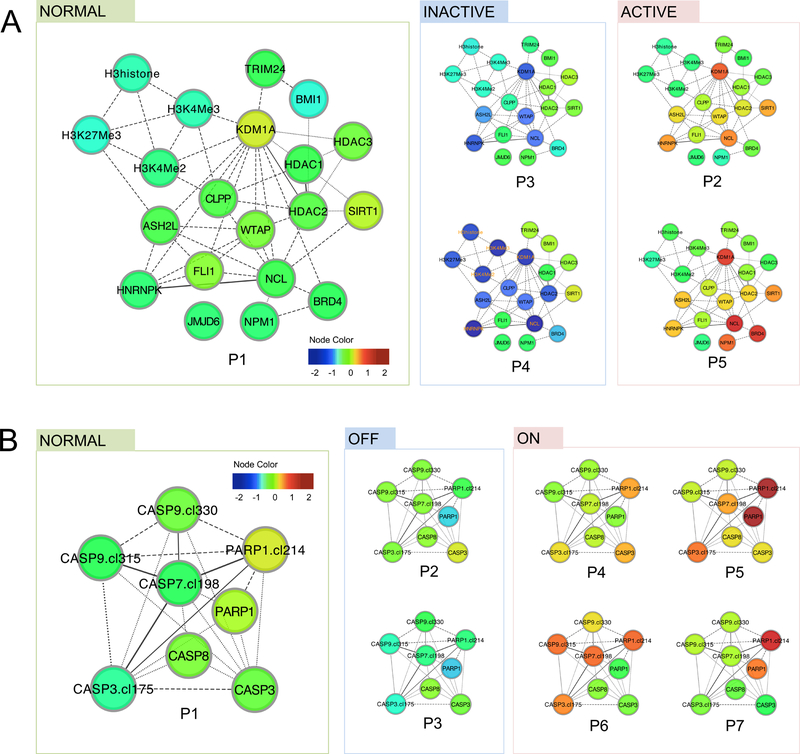
Figure 2. Functional patterns indicate varied functional states and alternative mechanisms.
Functional patterns were represented in protein networks, where the node color reflects the differential expression levels compared to the average expression levels in control samples. (A) In the Histone group, five functional patterns (P1-P5, ordered by similarity to patterns in control samples) were found. Specifically, P2 and P5 both result in an active state of histone modification (marked by high expression of histone regulators such as KDM1A and ASH2L), whereas P3 and P4 both result in an inactive state with reduced expression of these proteins. (B) In the Apoptosis-Occurring group, seven functional patterns were identified, where P4 to P7 all suggest heightened apoptosis activities (marked by higher levels of cleaved caspase 3, 7 and 9 as well as higher levels of cleaved PARP), despite their expression pattern differences.