Figure 4.
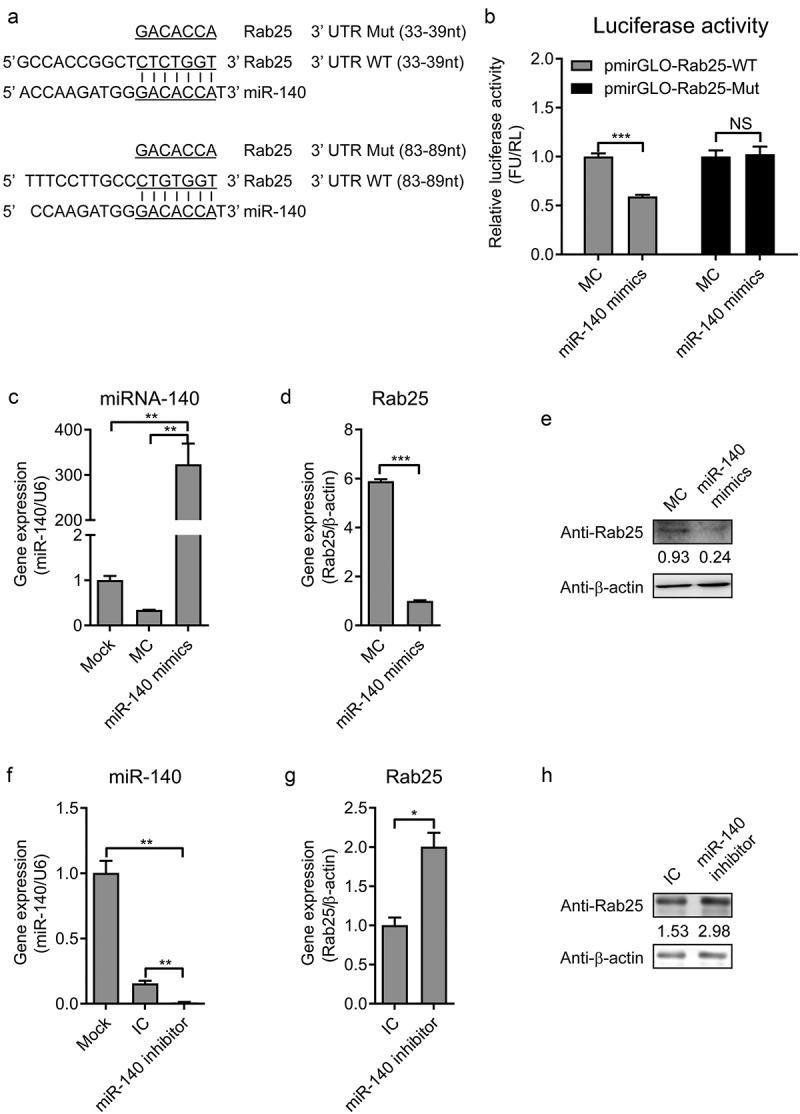
MiR-140 binds to the 3ʹ UTR of Rab25.
(a) RNA22 V2 and RNAhybrid online software were used to verify the target gene of miR-140. Alignments showed that there were two complementary sequences between miR-140 sequences and Rab25-3ʹ UTR sequences. Underlined sites indicate miR-140 seed region and mutated sequences are shown. (b) Dual-luciferase assays. Wild-type (WT) (pmirGLO-Rab25-WT) or mutant (pmirGLO-Rab25-Mut) luciferase-reporter vector and miR-140 mimics were co-transfected into cells. Luciferase activity were determined at 48 h post-transfection by dual-luciferase assays. (c, d, and e) 60 nM miR-140 mimics or mimics control (MC) were co-transfected into cells. (c) Real-time qRT-PCR analysis of miR-140 expression (normalized to U6 expression) at 48 h post-infection. (d) Real-time qRT-PCR analysis of Rab25 expression (normalized to β-actin expression) at 48 h post-infection. (e) Immunoblot analysis of Rab25 protein levels at 48 h post-infection. The number means the intensity of Rab25 protein level (top) normalized to that of β-actin. (f, g, and h) 60 nM miR-140 inhibitor or inhibitor control (IC) were co-transfected into cells. (f) Real-time qRT-PCR analysis of miR-140 expression (normalized to U6 expression) at 48 h post-infection. (g) Real-time qRT-PCR analysis of Rab25 expression (normalized to β-actin expression) at 48 h post-infection. (h) Immunoblot analysis of Rab25 protein levels at 48 h post-infection. The number means the intensity of Rab25 protein level (top) normalized to that of β-actin. Results in (a, c, d, f, and g) are expressed as mean ± SD of three independent experiments. p values were calculated using Student’s t-test. Western blots were analyzed and quantified using the Image J software. *P < 0.05, **P < 0.01, ***P < 0.001.
