Figure 1.
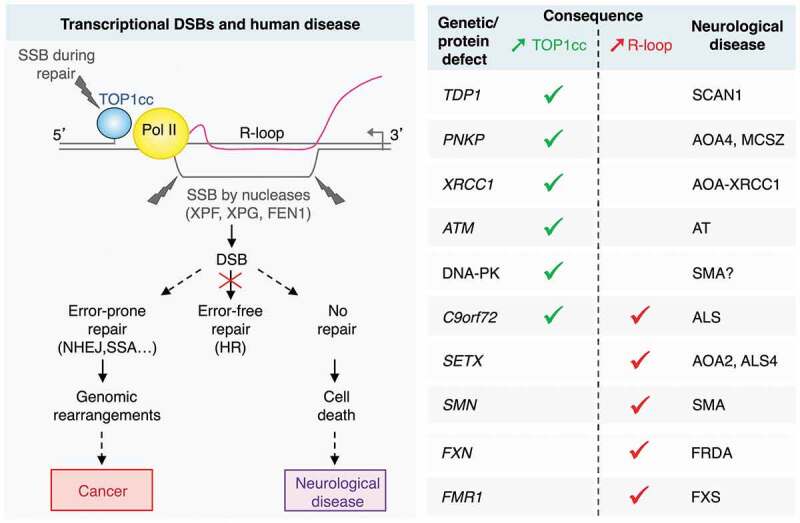
Outcomes of transcription-dependent DNA double-strand breaks (DSBs) in non-replicating cells. DSBs arise from two single-strand breaks (SSBs) on the opposing DNA strands: one SSB results from repair of a transcription-blocking topoisomerase I cleavage complex (TOP1cc), and the other from the cleavage of an R-loop by nucleases. In non-replicating cells, error-free homologous recombination (HR) is unavailable. Hence, DSBs could be repaired by error-prone pathways, such as non-homologous end joining (NHEJ) and single-strand annealing (SSA), which may promote genomic rearrangements and mutations. If left unrepaired, transcriptional DSBs may accumulate and induce cell death. The right-hand side table shows human neurological disorders, the genes (or protein expression) altered in these diseases and their association with increased TOP1ccs (green) or R-loops (red). AOA: ataxia with oculomotor apraxia; ALS: amyotrophic lateral sclerosis; AT: ataxia telangiectasia; ATM: ataxia telangiectasia mutated; C9orf72: chromosome 9 open reading frame 72; DNA-PK: DNA-dependent protein kinase; FEN1: flap structure-specific endonuclease 1; FRDA: Friedreich’s ataxia; FMR1: fragile X mental retardation 1; FXN: frataxin; FXS: fragile X syndrome; MCSZ: microcephaly, seizures, and developmental delay; PNKP: polynucleotide kinase 3ʹ-phosphatase; Pol II: RNA polymerase II, SCAN1: spinocerebellar ataxia with axonal neuropathy-1; SETX: senataxin; SMA: spinal muscular atrophy; SMN: survival motor neuron 1; TDP1: tyrosyl-DNA phosphodiesterase 1; XPF: xeroderma pigmentosum complementation group F; XPG: xeroderma pigmentosum complementation group G; XRCC1: X-ray repair cross-complementing 1.
