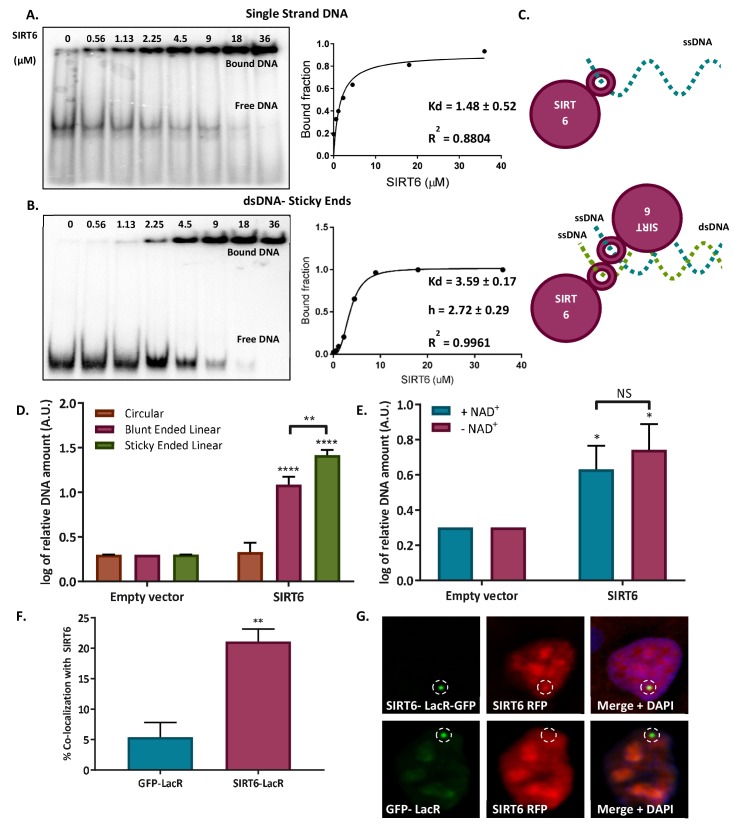
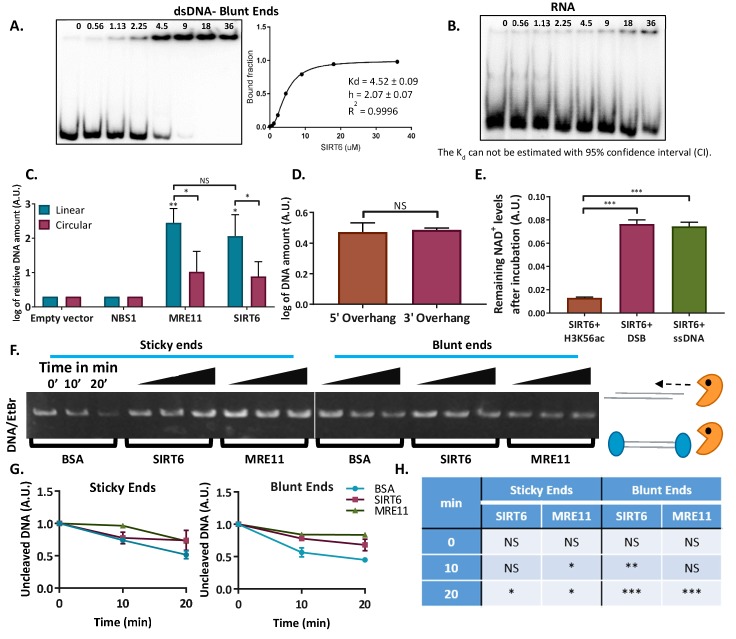
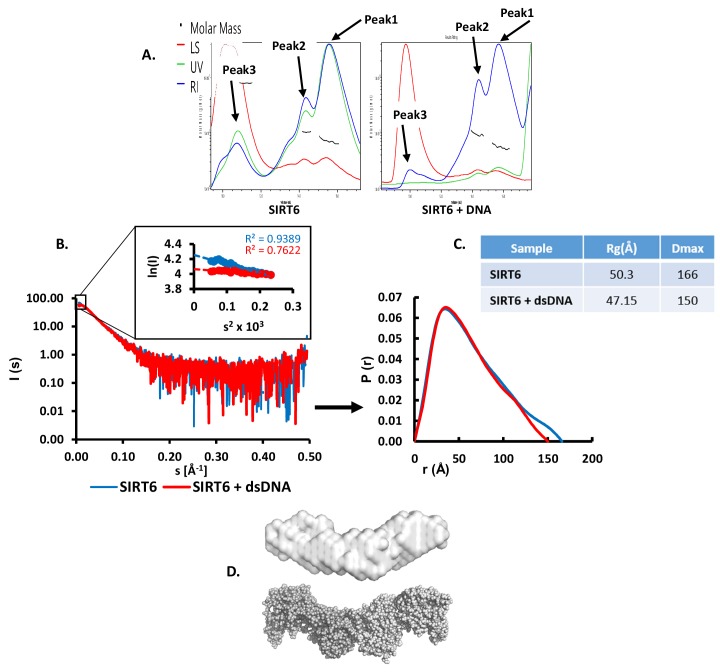
Figure 3. SIRT6 binds DNA with no intermediates.
(A–B) Gel retardation assay of 32 P-5' end-labeled single-strand DNAs and sticky ended dsDNAs as a function of increasing concentrations of SIRT6-His (ssDNA, Kd = 1.48 ± 0.52; sticky dsDNA, Kd = 3.59 ± 0.17). (C) Suggested model of SIRT6 binding to ssDNA as a monomer or open ssDNA ends of dsDNA as a dimer. (D) Ability of SIRT6-Flag to bind to the DNA of circular, blunt-ended and sticky-ended cleaved plasmids. The bar chart depicts averages for three replicate experiments (error bars show SEM), after logarithmic transformation. (E) SIRT6-Flag DNA-binding ability for an open-ended +plasmid +/- NAD. Data are averages from four experiments, with error bars representing SEM (after logarithmic transformation). (F, G) Dimerization of SIRT6 at the LacO site, represented by the recruitment of SIRT6-Cherry by SIRT6-LacR-GFP (n = 181, p<0.005) or GFP-LacR (n = 104). Data are averages from four experiments, with error bars representing SEM.