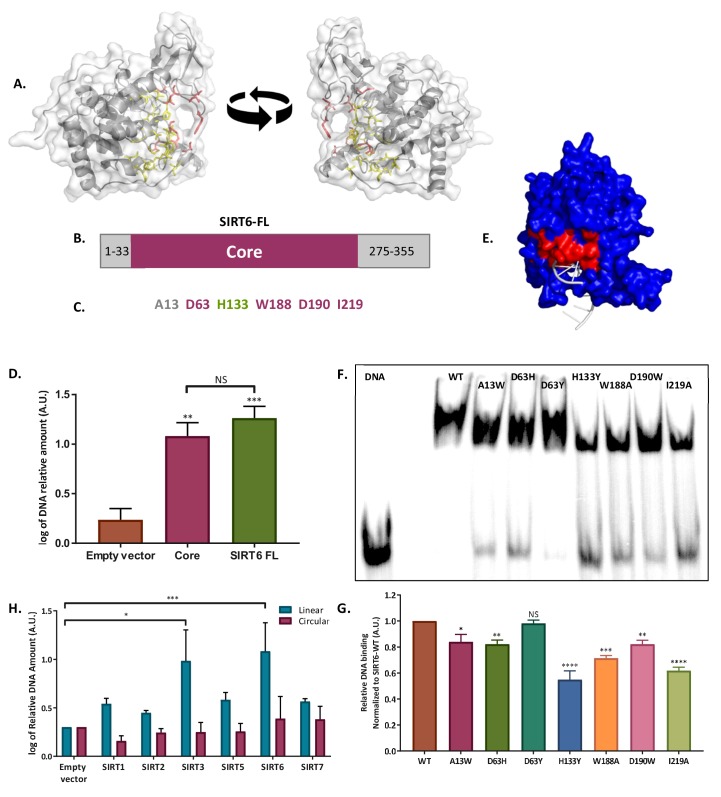
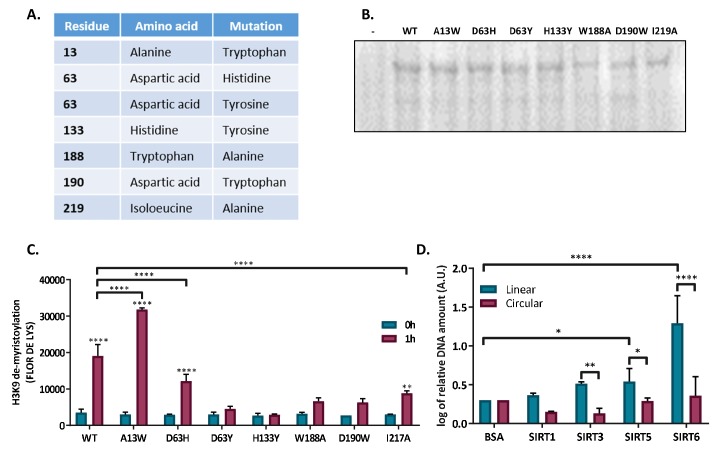
Figure 4. SIRT6 binds DSB through its core domain.
(A) Predicted DNA-binding site based on the published SIRT6 structure, (http://dnabind.szialab.org/). Highlighted in yellow are the predicted DNA-binding amino acids in the SIRT6 core domain; red highlights show the tunnel-forming amino acids that were mutated. (B) Schematic representation of the SIRT6 core domain. (C) List of amino acids that are predicted to participate in the ‘tunnel-like’ structure. (D) DNA binding of an open-ended plasmid by full-length SIRT6 (p<0.0005) and by the SIRT6-core domain (p<0.005). Data are the log of averages from three experiments (with error bars respresenting SEMs). (E) SIRT6 ssDNA-binding prediction, based on the known SIRT6 structure with bound ssDNA. (F, G) Gel retardation assay of 32 P-5′ end-labeled ssDNAs with SIRT6-MBP mutants. Data are averages from three experiments (with error bars representing SEMs). (H) Ability of Flag-tagged mammalian Sirtuins to bind the DNA of circular and linear plasmids. Data are averages from 4–7 experiments (with error bars representing SEMs) (*, p <0.05; **, p <0.005; ***, p <0.0005).