Fig. 5. Application for segmentation of immune cells.
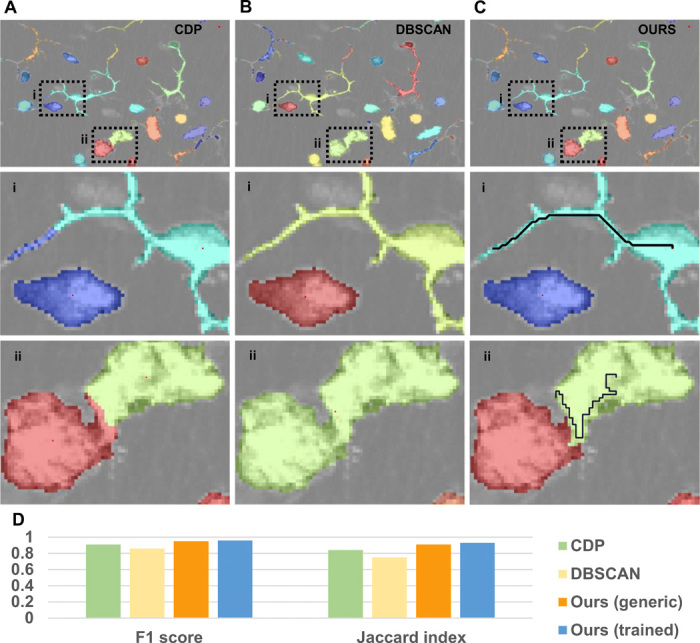
(A to C) Qualitative results (color-coded labels) produced by different density-based clustering algorithms for clustering pixels in space. Images show a confocal microscopy acquisition of murine immune cells, labeled with CD11c + GFP. (i) Magnification showing a cell with dendritic morphology close to a cell with globular shape. (ii) Magnification showing two cells with globular shape in contact. CDP (A) using a Euclidean metric correctly separates touching cells but associates a piece of dendrite to the wrong cell. DBSCAN (B) correctly reconstructs the shape of dendritic cells but is not able to separate touching cells with the same density-reachability criterion. The proposed method (C) correctly associates the dendrite of the dendritic cell and separates the touching cells. Black lines indicate the optimal path followed by the algorithm, from the cell centroid (density peak) to a point in the dendrite and in the touching region, respectively. (D) Quantitative performances. The F1 score and the Jaccard index are computed with respect to the ground truth. Here, the trained version of the algorithm achieved similar performances to the generic version. These are bounded because of the separation of cells with increased size into multiple subclusters.
