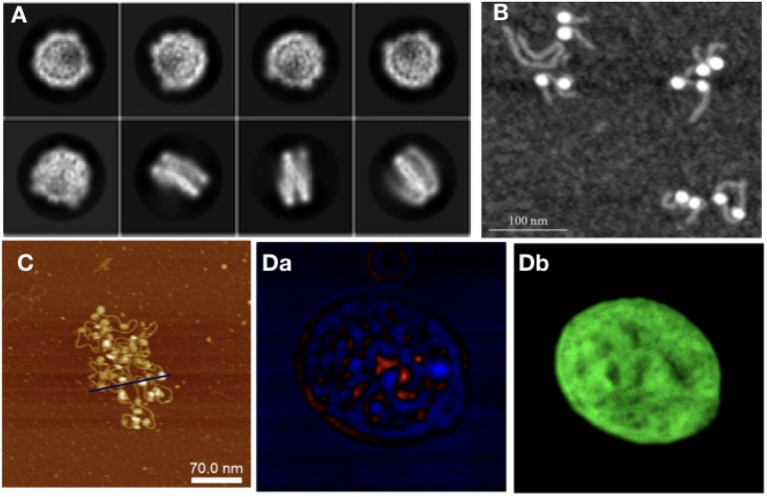
Figure 3.
As with modeling approaches, in experiments different techniques are required to study different orders of magnitude in chromatin: (A) NCP imaged with Cryo-Em (adapted from Kobayashi et al., 2019), (B) NCPs with histone tails AFM image (Filenko et al., 2012), (C) Nucleosome array, AFM image (adapted from Krzemien et al., 2017), (D) Isolated Hek nucleus imaged with CIDS (a), labeled with Hoechst for chromatin-DNA organization imaging. The fluorescence labeling (b) is used as a fingerprint of chromatin to demonstrate the correlation with the label-free approach using circular polarization excitation (Le Gratiet et al., 2018).