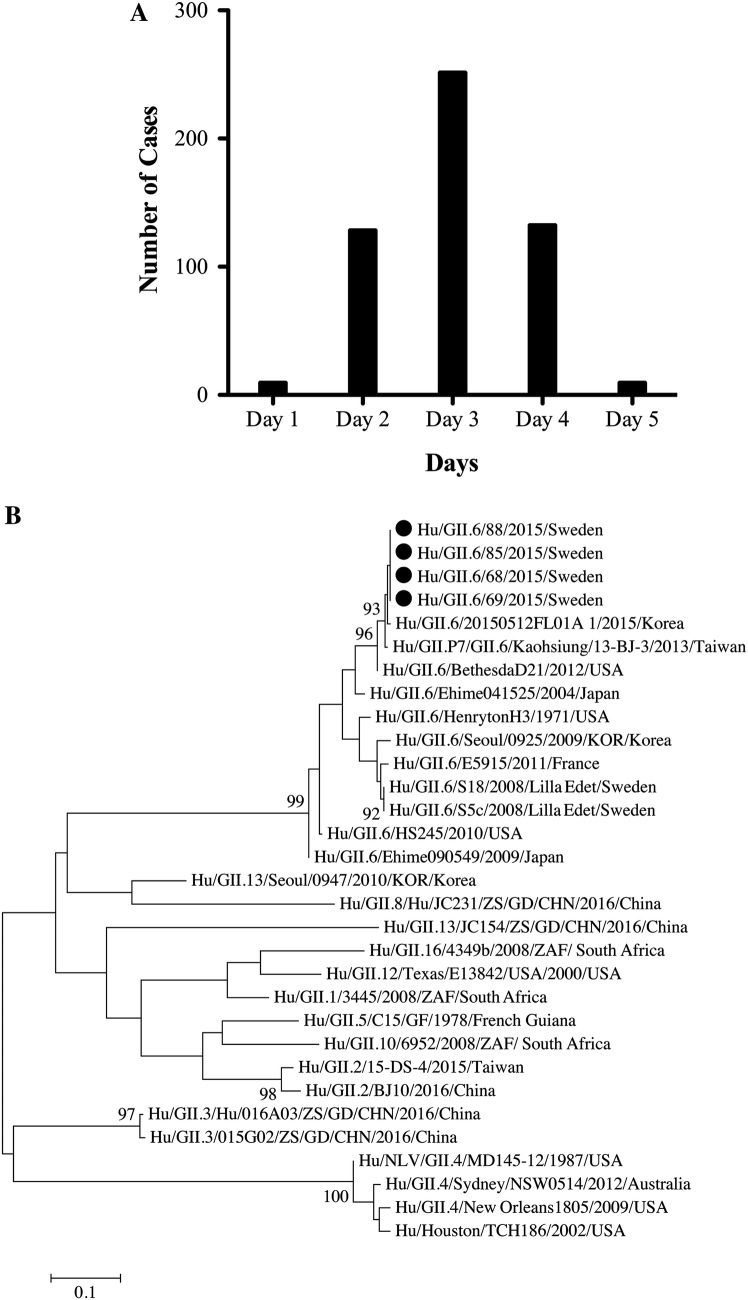
Fig. 1.
a Number of cases reported with norovirus-related illness (n = 542) during the first episode of GII.6 foodborne outbreak in a large office-based company in Stockholm, Sweden; b Phylogenetic tree showing clustering of four GII.6 norovirus strains (shown as filled circle) detected during the outbreak with reference GII.6 norovirus strains. The tree was generated on nucleotide sequences of the NS region (nt 1–272, ORF2). The scale bar represents number of nucleotide substitutions per site, and bootstrap values ≥ 85 are shown. Accession numbers of the strains sequenced are MH550088 for strain Hu/GII.6/68/2015/Sweden, MH550089 for strain Hu/GII.6/69/2015/Sweden, MH550090 for strain Hu/GII.6/85/2015/Sweden, and MH550091 for strain Hu/GII.6/88/2015/Sweden