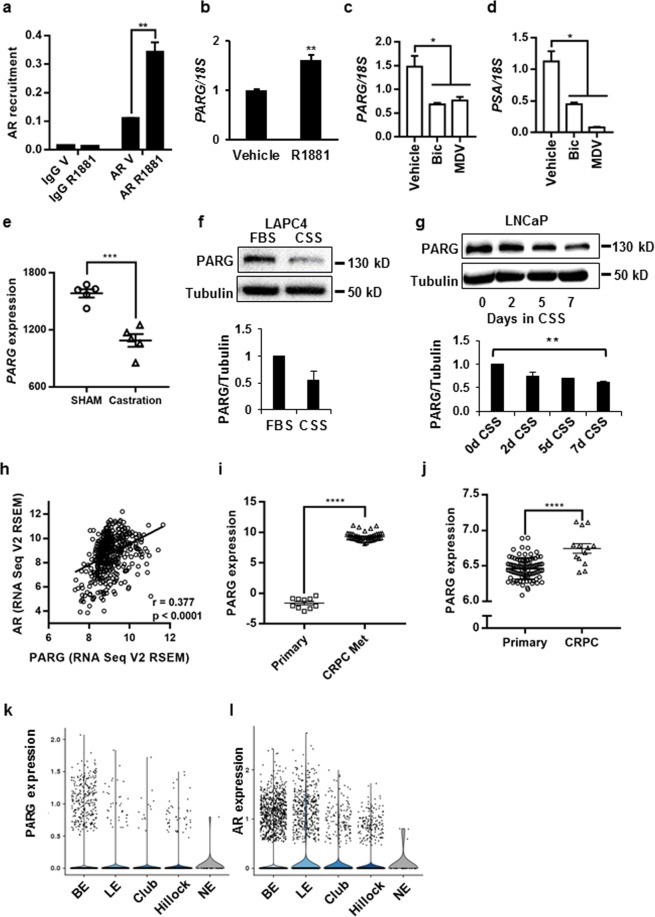
Figure 1.
AR regulation of PARG expression in independent models. (a) LNCaP cells were grown in medium supplemented with CSS and treated with ethanol vehicle or 10 nM of R1881 for 18 hours. AR recruitment to the PARG locus was compared by a ChIP Assay. Each bar represents an average of 3 biological replicates, p = 0.0012. The experiment was repeated 4 times. (b) RNA was extracted from LNCaP cells treated in parallel with (a), reverse transcribed, and used for qPCR to compare PARG mRNA levels. Expression was normalized to 18S. p = 0.00650. Each bar is an average of 3 biological replicates, the experiment was performed 4 times. (c,d) LNCaP cells were grown in medium supplemented with 10% FBS and treated with DMSO, 10–5 M of bicalutamide (Bic) or MDV3100 for 72 hours. RNA was isolated, reverse transcribed, and analyzed for PARG (c) and PSA (d) expression using 18S as control. Each bar is an average of 3 biological replicates. Experiment was repeated 3 times. (e) Values for PARG expression in LuCaP35 PDX samples grown in intact (N = 5) and castrated (N = 5) mice were exported from GSE33316 and averaged (p = 0.00025). (f) Top: PARG expression in LAPC4 cells grown in FBS or CSS supplemented media for 48 hours. Representative western blot shown. Bottom: average quantification of PARG protein levels normalized for Tubulin from 3 independent experiments. (g) LNCaP cells grown in FBS or CSS for 2, 5, and 7 days. Experiments f-g were repeated 3 times and representative blots are shown. (h) Correlation between PARG expression and AR expression in 499 prostate adenocarcinoma samples (TCGA, Provisional). (i) PARG expression in samples of 10 primary prostate tumors and 45 metastatic CRPC tumors in data set GSE74367. (j) PARG expression in 94 primary prostate tumors and 13 CRPC prostate tumors in data set GSE70770. (k,l) The violin plots show the expression of PARG (k) and AR (l) in basal epithelia (BE), luminal epithelia (LE), club epithelia (Club), Hillock epithelia (Hillock), and neuroendocrine (NE) of human prostate cells (GSE120716, http://strandlab.net/analysis.php). (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001).