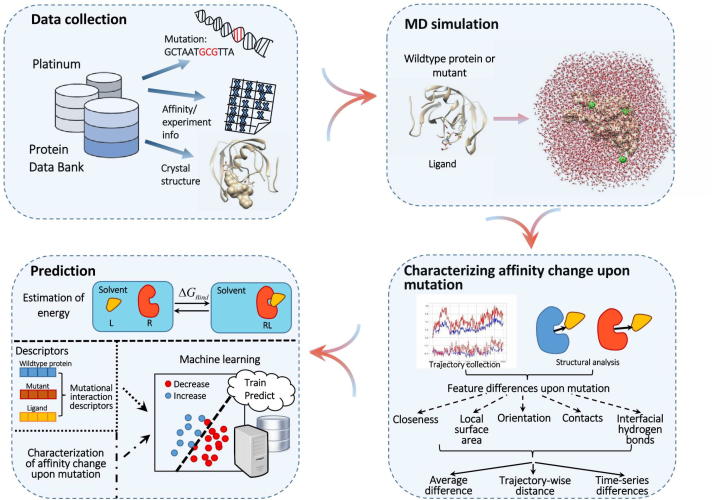
Fig. 1.
Overall framework for predicting the impacts of mutations on protein-ligand binding affinity. In the data-collection phase, mutation, affinity (ligand-binding affinity measurements for each pair of wild-type protein (WTP) and its mutant), experimental (experiment conditions for deriving the ligand-binding affinity) and structural (crystal structures of WTP-ligand and mutant-ligand complexes) data were conjunctionally collected from Platinum and Protein Data Bank. The molecular dynamics (MD) simulation step involved each WTP-ligand or mutant-ligand system, and adopted the explicit-solvent model. Next, the trajectory frames for each system were collected, and the difference between each pair of WTP-ligand and mutant-ligand systems was quantified according to several local geometrical features (closeness, local surface area, orientation, contacts and interfacial hydrogen bonds) in these frames. Finally, in the prediction phase we adopted machine-learning methods to relate such feature differences to the mutation impact on protein-ligand binding affinity. Prediction based on direct estimation of binding free energy and that based on molecular descriptors were also implemented as benchmarks.