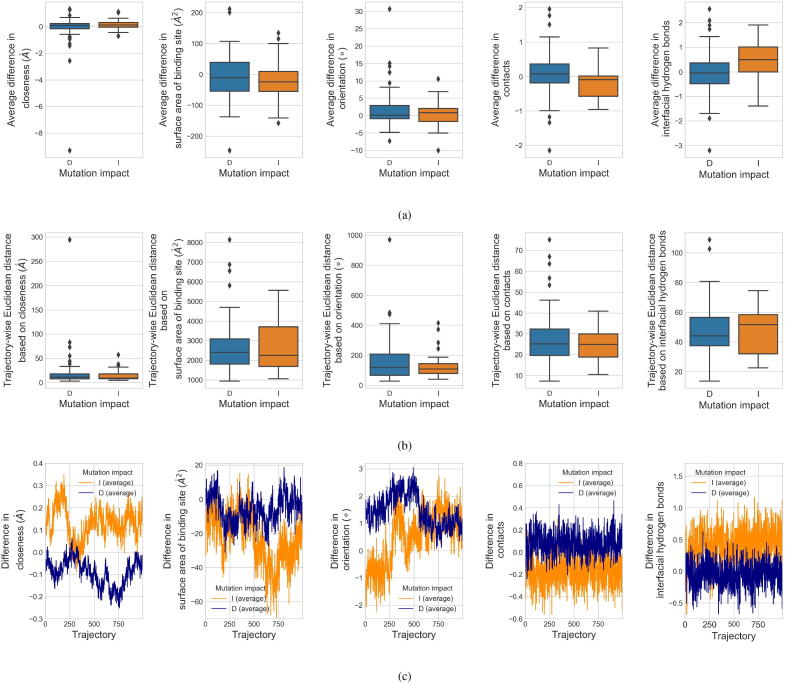
Fig. 5.
To characterize the affinity change upon mutation, difference between each pair of WTP-ligand and mutant-ligand systems was quantified by local geometrical features (closeness, surface area of binding site, orientation, contacts and interfacial hydrogen bonds) extracted from the molecular dynamics (MD) trajectories of the systems. Feature distributions for the two mutation impacts, decreased affinity (D) and increased affinity (I), are shown. Panel (a) shows the differences in each feature averaged over all the MD structural snapshots for each mutation. Panel (b) shows the trajectory-wise Euclidean distance based on each feature for the mutations. Panel (c) shows the time-vary differences in each feature during the 2 ns-simulation, averaged in each mutation group.