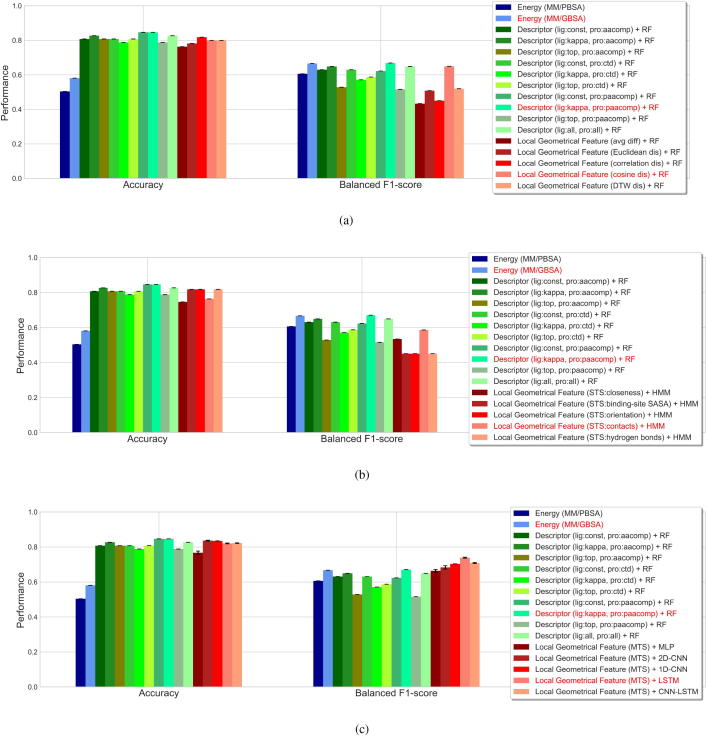
Fig. 6.
Performance evaluation of predicting mutation impacts on protein-ligand binding affinity. Performance was evaluated based on the overall accuracy and balanced F1 score. Results from the estimation of binding-free-energy difference and from the prediction based on molecular descriptors are provided as benchmarks. In the energy-based predictions, MM/PBSA and MM/GBSA methods were employed for estimating the binding free energy. In the descriptor-based predictions, generally used descriptors for the ligands (Constitutional descriptors (const), Kappa shape indices (kappa) and Topological descriptors (top)) and proteins (Amino acid composition descriptors (aacomp), Composition, translation and distribution descriptors (ctd) and Type 1 pseudo amino acid composition descriptors (paacomp)) were adopted. On top of these descriptors, the mutational interaction descriptors were constructed by concatenations or tensor products, and used for the prediction of mutation impact by random forests (RFs). Our method characterized protein-ligand binding affinity change upon mutation by several local geometrical features (closeness, solvent accessible surface area (SASA) of binding site, orientation, contacts and interfacial hydrogen bonds) and monitored such feature differences upon mutation in the molecular dynamics (MD) simulations, after which a number of machine-learning methods were adopted for prediction of mutation impact. (a) RF-based predictions according to the features extracted as the average differences or trajectory-wise distances between each WTP-ligand and mutant-ligand systems. Distance metrics of the Euclidean, Pearson correlation, cosine and dynamic time warping (DTW) were considered. (b) Hidden Markov models (HMMs)-based predictions according to single time-series (STS) features, extracted as the time-varying differences in each of the five local geometrical features. (c) Prediction according to multiple time-series (MTS) features (based on all of the five local geometrical features) by shallow or deep neural networks. These neural networks include multilayer perceptron (MLP), convolutional neural networks (CNNs) with 2D convolutional layers, CNNs with 1D convolutional layers, recurrent neural networks (RNNs) with long short-term memory (LSTM) cells, and neural networks composed of CNNs and LSTM (CNN-LSTM).