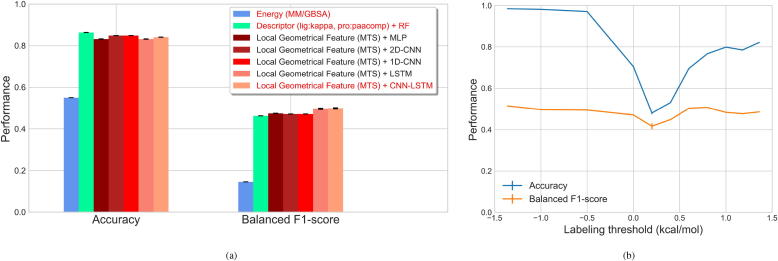
Fig. 10.
Performance evaluation of predicting mutation impacts on protein-ligand binding affinity for the Abl-mut data set. Performance was evaluated based on the overall accuracy and balanced F1 score. (a) Results of our method, which uses machine-learning techniques to monitor the time-varying local geometrical feature differences upon mutation in the molecular dynamics (MD) simulations, are displayed. Shallow or deep neural networks include multilayer perceptron (MLP), convolutional neural networks (CNNs) with 2D convolutional layers, CNNs with 1D convolutional layers, recurrent neural networks (RNNs) with long short-term memory (LSTM) cells, and neural networks composed of CNNs and LSTM (CNN-LSTM) were used. Results from the estimation of binding-free-energy difference (MM/GBSA) and from the prediction based on molecular descriptors (Kappa shape indices (kappa) for ligands and Type 1 pseudo amino acid composition descriptors (paacomp) for proteins, and random forests (RFs) for predictions) are provided for comparisons. (b) Evaluation of the effects of different labeling cutoffs on the prediction performance. Prediction performance was evaluated based on LSTM.