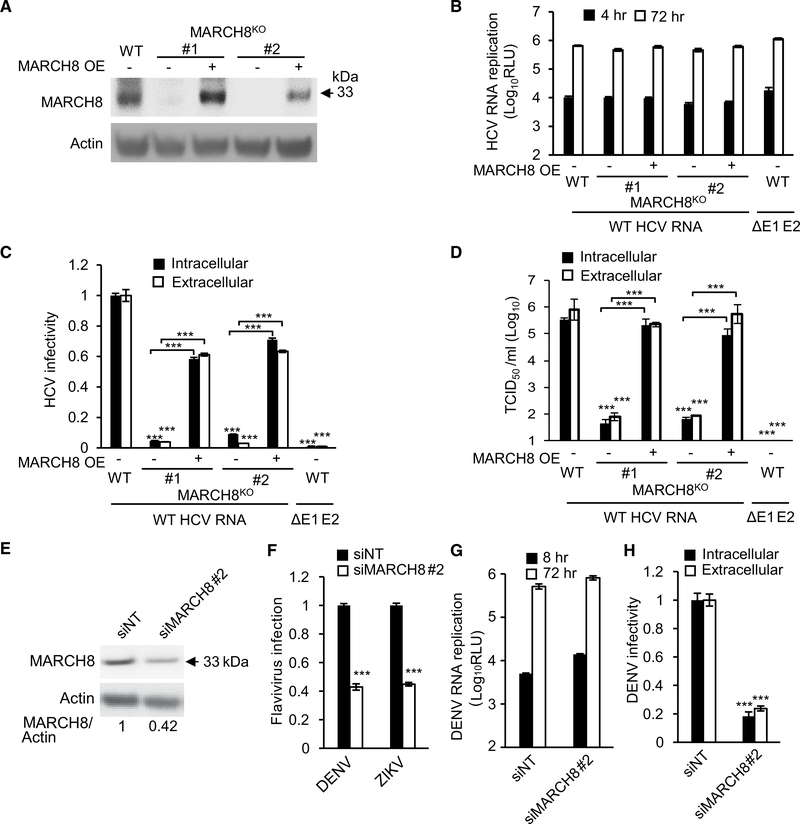
Figure 5. MARCH8 Is Required for HCV and DENV Assembly.
(A) MARCH8 protein by western blot in control Huh7.5.1 (WT) cells, two cell lines deleted for MARCH8 via CRISPR/Cas9, and these MARCH8KO cell lines upon ectopic expression of MARCH8.
(B) HCV RNA replication in these cells 6 and 72 hr after electroporation with WT HCV RNA or E1-E2 glycoprotein-deleted assembly defective HCV mutant (ΔE1–E2), measured by luciferase assays (RLU, relative light units; OE, overexpression).
(C) HCV infectivity measured via luciferase assays by inoculating naive cells with lysates (intracellular) and supernatants (extracellular) derived from electroporated cells 72 hr after electroporation.
(D) Intracellular and extracellular viral titers measured by limiting dilution assays. TCID50, 50% tissue culture infectious dose.
(E) MARCH8 protein in Huh7 cells transfected with the indicated siRNAs (numbers represent MARCH8-to-actin protein ratio relative to the NT control).
(F) DENV2 (MOI = 0.1) and ZIKV (MOI = 0.05) infection in MARCH8-depleted cells, measured by luciferase assays at 48 and 72 hr, respectively, and normalized to cell viability.
(G) DENV2 RNA replication in MARCH8-depleted Huh7 cells measured by luciferase assays at 8 and 72 hr after electroporation with DENV RNA.
(H) DENV2 infectivity measured via luciferase assays by inoculating naive cells with lysates (intracellular) and supernatants (extracellular) from electroporated cells.
(B)–(D) represent data pooled from three independent experiments each with 3–6 biological replicates. (F)–(H) are representative experiments out of two conducted. Shown are means ± SD; ***p < 0.001 relative to corresponding WT or NT controls by one-way (C, D, F, and H) or two-way (B and G) ANOVA with Dunnett’s (F, G, and H) or Tukey’s (B, C, and D) post hoc tests.