Fig. 5.
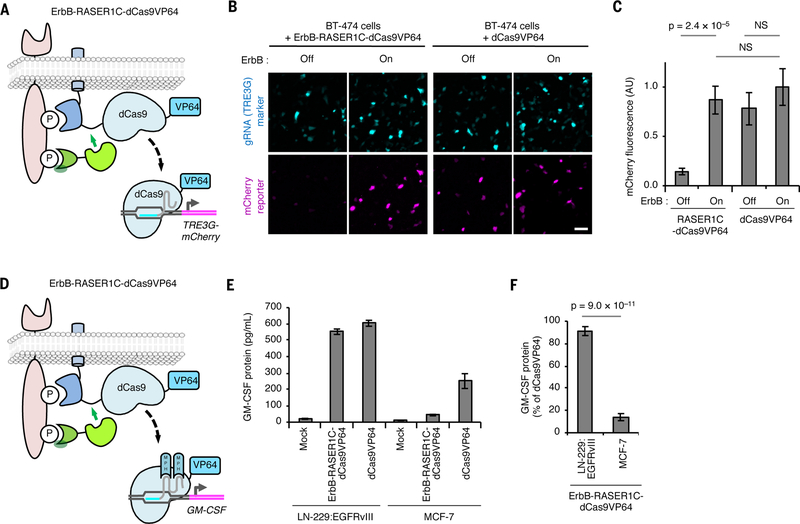
RASER can be programmed to activate endogenous genes of choice selectively in ErbB-driven cancer cells. (A) Schematic of the ErbB-RASER1C-dCas9VP64 system regulating a reporter gene. dCas9 with a VP64 transcriptional activation domain will be released in the presence of constitutively active ErbB to activate TRE3G-mCherry. (B) ErbB-RASER1C-dCas9VP64 and a TRE3G-directed sgRNA activate TRE3G-mCherry in an ErbB activity-dependent manner in BT-474 cells. mTurquoise is marker for gRNA expression. Scale bar, 100 μm. (C) Quantification of (B). AU, arbitrary units. NS, not significant. Differences between conditions were assessed by the Kruskal-Wallis test followed by two-tailed Dunn’s posthoc tests. p = 6.5 × 10–8 for the overall null hypothesis of no difference between groups (n = 50 randomly selected transfected cells). (D) Schematic of the ErbB-RASER1C-dCas9VP64 system regulating endogenous GM-CSF. (E) RASER activates endogenous GM-CSF in ErbB-hyperactive cells. LN-229:EGFRvIII cells expressing constitutively active ErbB1 and MCF-7 cells with normal ErbB were transfected with ErbB-RASER1C-dCas9VP64, GM-GSF gRNA containing MS2-binding sequences, and MS2-p65-HSF1 (MPH). After 24 h, released GM-CSF was quantified by ELISA (n = 3 biological replicates). (F) Induction of GM-CSF protein by RASER1C-dCas9VP64 is > 4-fold more efficient in LN-229:EGFRvIII cells than in ErbB-normal MCF-7 cells (n = 3 biological replicates). All error bars represent s.e.m.