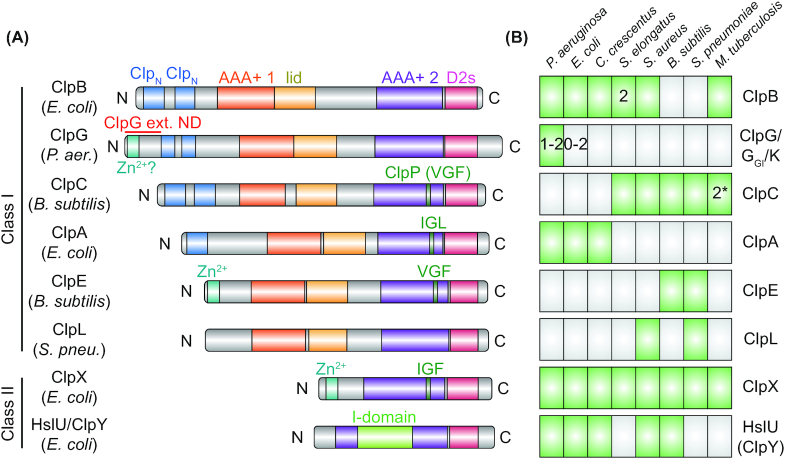
Figure 3.
Distribution of Hsp100 proteins among bacteria and Hsp100 domain structure. (A), Domain organization of bacterial Hsp100 homologs from different bacterial species. Domains annotated using InterPro (http://www.ebi.ac.uk/interpro/) show the respective identifiers. Hsp100 proteins can be categorized as two classes according to the number of AAA+ domains they contain: Class I Hsp100s ClpA, ClpB, ClpC, ClpE, GlpG (or ClpK) and ClpL have two while class II Hsp100s ClpX and HslU (also called ClpY) have only one. The AAA+ core domains (AAA+ 1/2; IPR003959) are C-terminally bordered by the AAA+ lid domain (lid; IPR041546) or the D2 small domain (D2s; IPR019489), respectively. HslU (ClpY) has an intermediate domain (I-domain, residues 108–243) inserted into its AAA+ core domain. The remaining domain organization of Hsp100s is protein specific, with differences in the length of the region between the AAA+ domains as well as in the N-termini, which may contain one or several conserved Clp N-terminal domains (ClpN; IPR004176) or a zinc-binding motif (Zn2+; IPR010603 in ClpX). The stand-alone disaggregases ClpG and ClpGGI possess an N-terminal extension with a putative zinc-binding motif (Zn2+?), which is involved in protein aggregate interaction (ClpG), or ATPase activity regulation (ClpGGI). HslU (ClpY) associates with the peptidase HslV (ClpQ) while ClpA, ClpC, ClpE and ClpX associate with the peptidase ClpP. ClpP interaction involves a tripeptide motif interaction loop in an AAA+ domain of the Hsp100s (tripeptide sequence indicated). ClpL lacks a known ClpP interacting motif and it is unknown if it interacts with a peptidase subunit. (B), Distribution of Hsp100 proteins in selected bacterial species belonging to the Proteobacteria (Pseudomonas aeruginosa, Escherichia coli and Caulobacter crescentus), Cyanobacteria (Synechococcus elongatus), Firmicutes (Staphylococcus aureus, Bacillus subtilis and Streptococcus pneumoniae) and Actinobacteria (Mycobacterium tuberculosis). ClpG belongs to the Pseudomonas aeruginosa core genome but a second homolog (ClpGGI) can be found on genomic island 1 of P. aeruginosa clone C. Most E. coli species lack ClpG, however, some E. coli strains harbor a ClpG or ClpGGIhomolog on a genomic island or on mobile genetic elements. Synechococcus elongatus possesses two ClpB homologs (ClpB1 and ClpB2) and two potential ClpC homologs (ClpC1 and the unusual truncated ClpC2/ClpX’) can be found in Mycobacterium tuberculosis.