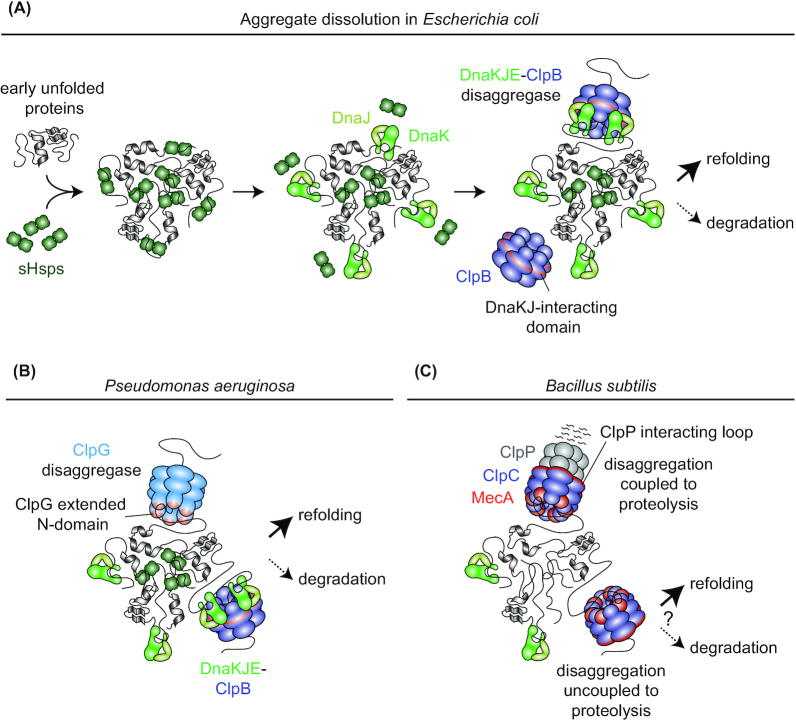
Figure 4.
Protein disaggregation in different bacterial species. (A), Protein disaggregation via the DnaKJE-ClpB bichaperone machinery in E. coli (GrpE not shown). sHSPs (IbpA and IbpB in E. coli) bind recently unfolded proteins and are incorporated into aggregates while maintaining proteins in a near-native state that facilitates eventual disaggregation by DnaKJE-ClpB. Together with the co-chaperone DnaJ, DnaK binds to aggregated protein and displaces sHSPs from the aggregate surface. DnaK recruits the ClpB disaggregase to the aggregate and this interaction stimulates high ClpB ATPase activity. Substrates are threaded through the central pore of ClpB and, after extraction from the aggregate, can be refolded (preferred) or degraded. (B), Protein disaggregation machineries in Pseudomonas aeruginosa. In addition to the DnaKJE-ClpB disaggregation machinery, P. aeruginosa employs ClpG which does not require stimulating factors for high disaggregation activity. Proteins extracted by both machineries can subsequently be refolded or degraded. (C), Potential protein disaggregation mechanisms in B. subtilis. B. subtilis lacks ClpB or another stand-alone disaggregase. Instead, the ClpP interacting loop containing Hsp100 ClpC interacts with the adaptor MecA and binds to aggregated proteins to drive their disaggregation in vitro. Although protein degradation-coupled disaggregation by MecA-ClpCP has been shown to be more efficient, MecA-ClpC can also disaggregate proteins without associating with ClpP. Thus, potential disaggregation through MecA-ClpC not associated with ClpP could allow for the refolding (or the downstream degradation by another protease) of extracted proteins in vivo.