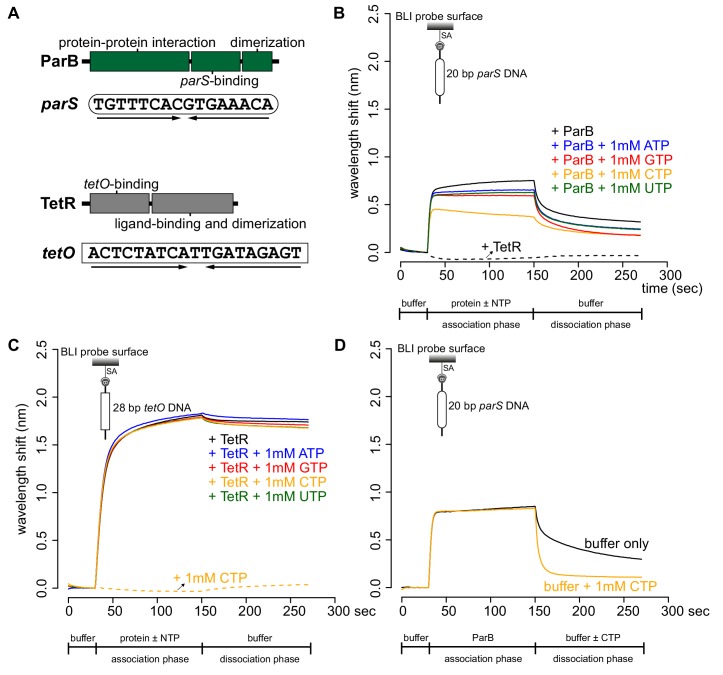
Figure 1. CTP reduces the nucleation of Caulobacter ParB at parS.
(A) The domain architecture of ParB (dark green) and TetR (grey), and their respective DNA-binding sites parS and tetO. Convergent arrows below DNA-binding sites indicate that parS and tetO are palindromic. (B) Bio-layer interferometric (BLI) analysis of the interaction between a premix of 1 μM ParB-His6 dimer ± 1 mM NTP and a 20 bp DNA duplex containing parS. Biotinylated DNA fragments were immobilized onto the surface of a Streptavidin (SA)-coated probe (See Materials and methods). The BLI probe was dipped into a buffer only solution (0–30 s), then to a premix of protein ± NTP (30–150 s: association phase), and finally returned to a buffer only solution (150–270 s: dissociation phase). Sensorgrams were recorded over time. BLI analysis of the interaction between 1 μM TetR-His6 and a 20 bp parS probe was also recorded (a negative control). (C) BLI analysis of the interaction between a premix of 1 μM TetR-His6 ± 1 mM NTP and a 28 bp DNA duplex containing tetO. BLI analysis of the interaction between 1 mM CTP and a 28 bp tetO probe was also recorded (a negative control). (D) BLI analysis of the interaction between 1 μM Caulobacter ParB-His6 (without CTP) and a 20 bp parS DNA. For the dissociation phase, the probe was returned to a buffer only or buffer supplemented with 1 mM CTP. All buffers used for experiments in this figure contained Mg2+. Each BLI experiment was triplicated and a representative sensorgram was presented.
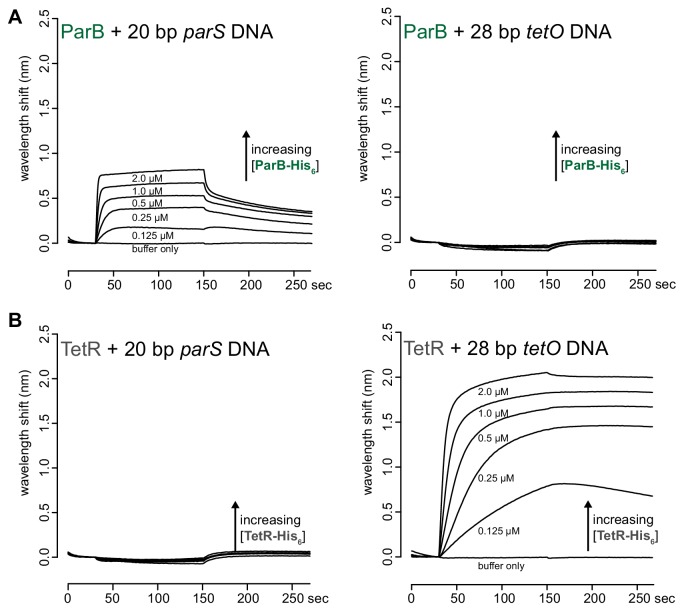
Figure 1—figure supplement 1. ParB and TetR bind specifically to their cognate-binding sites parS and tetO, respectively.
Figure 1—figure supplement 2. BLI analysis of the interaction between purified Caulobacter ParB and NTP in buffers lacking Mg2+.

Figure 1—figure supplement 3. BLI analysis of the interaction between purified Caulobacter ParB and cytidine mono-, di-, or triphosphate.