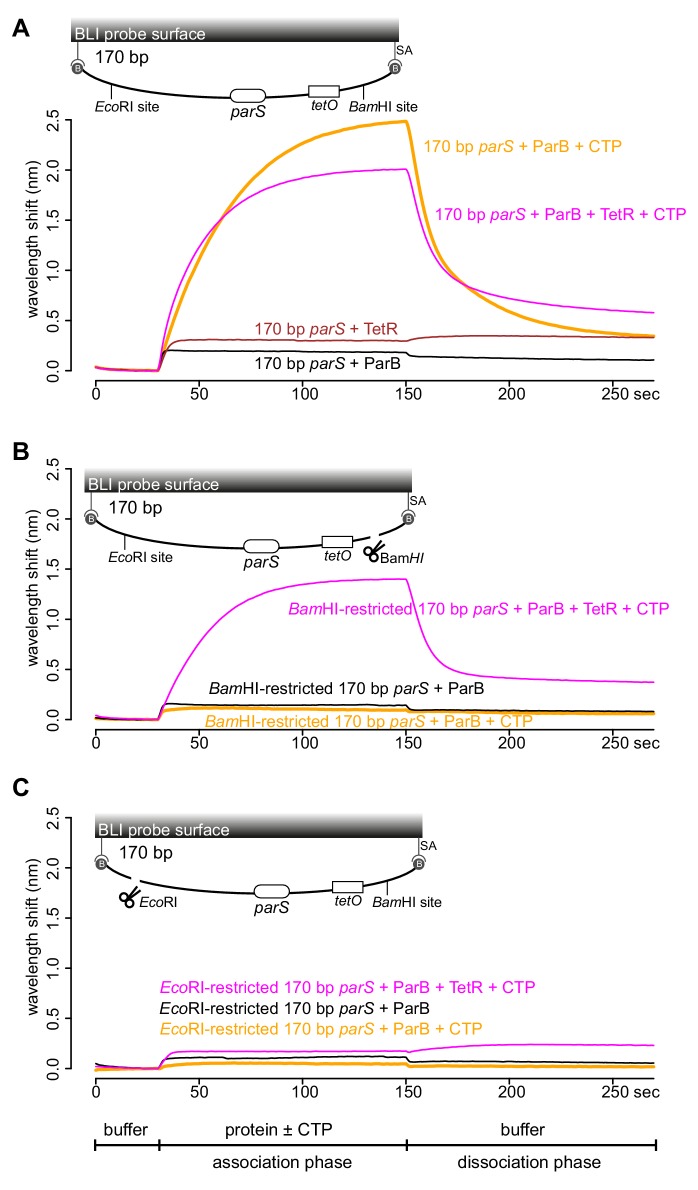
Figure 4. TetR-tetO binding restores ParB association with an open DNA substrate.
(A) BLI analysis of the interaction between a premix of 1 μM Caulobacter ParB-His6 ± 1 mM CTP ± 1 μM TetR-His6 and a 170 bp dual biotin-labeled DNA containing a parS site. (B) Same as panel A but immobilized DNA fragments have been restricted with BamHI before BLI analysis. (C) Same as panel A but immobilized DNA fragments have been restricted with EcoRI before BLI analysis. Schematic of DNA fragments together with the relative positions of parS, tetO, and restriction enzyme recognition sites are shown above the sensorgram. Each BLI experiment was triplicated and a representative sensorgram was presented. All buffers used for experiments in this figure contained Mg2+.