The azoxymethane carcinogen model of non-familial colorectal cancer has been used in mice to identify six new susceptibility loci and confirm 18 of 24 previous detected susceptibility loci. Using a population-based approach, the genetic architecture of colon cancer...
Keywords: cancer susceptibility, mouse models, heterogeneity, genetic architecture
Abstract
The azoxymethane model of colorectal cancer (CRC) was used to gain insights into the genetic heterogeneity of nonfamilial CRC. We observed significant differences in susceptibility parameters across 40 mouse inbred strains, with 6 new and 18 of 24 previously identified mouse CRC modifier alleles detected using genome-wide association analysis. Tumor incidence varied in F1 as well as intercrosses and backcrosses between resistant and susceptible strains. Analysis of inheritance patterns indicates that resistance to CRC development is inherited as a dominant characteristic genome-wide, and that susceptibility appears to occur in individuals lacking a large-effect, or sufficient numbers of small-effect, polygenic resistance alleles. Our results suggest a new polygenic model for inheritance of nonfamilial CRC, and that genetic studies in humans aimed at identifying individuals with elevated susceptibility should be pursued through the lens of absence of dominant resistance alleles rather than for the presence of susceptibility alleles.
COLORECTAL cancer (CRC) is the second leading cause of cancer-related death in the United States, with over 145,000 new cases diagnosed each year (2019). Although a small fraction of these cases are due to well-characterized hereditary syndromes such as Familial Adenomatous Polyposis (FAP) and Hereditary Non-Polyposis Colon Cancer (HNPCC), the vast majority of CRCs are considered sporadic or nonfamilial (Burt et al. 1992). However, numerous studies indicate that nonfamilial CRC is the result of the interaction among multiple, low penetrance (small effect) alleles and environmental factors (Hutter et al. 2012; Montazeri et al. 2016; Liu et al. 2019; Yang et al. 2019b).
The discovery that dimethylhydrazine (DMH) and its metabolite azoxymethane (AOM) are colon-specific carcinogens paved the way to model nonfamilial CRC in rodents (Druckrey et al. 1967). Using a variety of dose regimes, it was shown that inbred mouse strains vary extensively in their susceptibility to these carcinogens, mirroring variable susceptibility to CRC thought to exist among humans. In contrast to genetic models such as ApcMin or deficiency for Smad3 (Moser et al. 1990; Zhu et al. 1998), tumors developing in AOM-treated mice arise almost exclusively in the distal colon. AOM-induced tumors are also molecularly similar to nonfamilial human CRCs, showing activation of the WNT/CTNNB1 (beta-catenin) signaling pathway and upregulation of Myc and Ccnd1 (Tulchin et al. 1988; Wang et al. 1998; Suzui et al. 1999; Kaiser et al. 2007). Because of the similarities with nonfamilial CRC in humans, the AOM model has been used to provide insights into molecular pathways associated with cancer development (Takahashi and Wakabayashi 2004; Chen and Huang 2009), to test chemopreventative and chemotherapeutic approaches (Reddy 2004; Waly et al. 2014; Manna et al. 2015; Odun-Ayo et al. 2015; Pedro et al. 2016; Bi et al. 2017; Md Nasir et al. 2017; Wu et al. 2017), and to identify genetic (Ruivenkamp et al. 2003; Meunier et al. 2010, 2011, 2013; Eversley et al. 2012; Liu et al. 2012) and environmental (Bissahoyo et al. 2005; Takahashi et al. 2013; Piazzi et al. 2019) factors that contribute to nonfamilial CRC susceptibility .
Despite the extensive use of the AOM model for experimental cancer research, few studies have evaluated the relative susceptibility to AOM-induced CRC (Nambiar et al. 2003; Ruivenkamp et al. 2003; Meunier et al. 2010, 2011, 2013; Liu et al. 2012), predominantly using mice derived from commonly used strains that represent a limited pool of ancestors (Yang et al. 2007). Even then, these studies used only a few mice per strain, which greatly reduces the accuracy of measuring strain response. The genetic variability known to be present in mice suggests that a population of mouse strains representing diverse origins is an excellent model for the heterogeneous human population (Harrill et al. 2009). However, the variability in response to carcinogens, even using identical mice within an inbred strain, can greatly limit accuracy of measuring inbred line responses and thus reduces the power to identify cancer susceptibility modifiers. In the present study, an extensive population-level characterization of response to AOM-carcinogenesis was performed. This study demonstrates extensive variability in susceptibility to AOM-induced CRCs across mouse strains. Similar to genome-wide association studies (GWAS) in humans (Tomlinson et al. 2007, 2008; Zanke et al. 2007; COGENT Study 2008; Tenesa et al. 2008; Tenesa and Dunlop 2009; Theodoratou et al. 2012; Schumacher et al. 2015; Montazeri et al. 2016; Tanikawa et al. 2018; Bien et al. 2019; Lu et al. 2019), genome-wide association analysis using mouse strains reveals susceptibility to be highly polygenic, with few large-effect susceptibility alleles and different modifier alleles influencing different aspects of carcinogenesis (Liu et al. 2012). Additionally, the genetic architecture of susceptibility to AOM-induced CRC was investigated using crosses between strains with varying susceptibilities, which demonstrated that genome-wide resistance to CRC is dominant. The data indicate a new model for CRC susceptibility, where individuals at high risk for developing nonfamilial CRC lack sufficient numbers of small-effect, dominant resistance alleles, rather than having specific susceptibility alleles.
Materials and Methods
Mouse strains and husbandry
Mice were obtained from The Jackson Laboratory or Taconic, and bred inhouse. Mice were group housed, except for individually housed SJL/J and NZB/B1NJ males, in microisolators or ventilated racks, at constant temperature and humidity on a 12-hr light/12-hr dark in a pathogen-free barrier facility negative for Helicobacter sp. Crosses between select strains were performed to produce F1 hybrids, F2 intercross, and N2 backcross progeny. Mice were provided with LabDiet 5010 and autoclaved water ad libitum. The studies were approved by the Institutional Animal Care and Use Committee.
Carcinogen treatment and tumor characterization
Between 2 and 4 months of age, mice were given 4-weekly intraperitoneal injections of AOM at 10 mg/kg body weight, diluted in phosphate buffered saline (PBS), a dosing previously shown to maximize interstrain differences in susceptibility (Bissahoyo et al. 2005). All AOM used in the study was from a single lot (Sigma). Mice, except for KK/H1J, were killed by CO2 asphyxiation 6 months after the first AOM dose. KK/H1J mice were killed after 5 months because of intestinal blockage caused by the development of large tumors. Age-matched controls for C57BL/6J, DBA/2J, and SWR/J mice were injected with PBS. No control mice developed intestinal tumors.
Upon euthanasia, colons were dissected, gently flushed with PBS, and splayed open along their longitudinal axis. Tumors 1 mm or larger in diameter were counted under a dissecting microscope, their diameters measured, and locations along the length of the colon recorded. No tumors were detected in the small intestine.
Association mapping
Two methods of association mapping were performed to identify regions harboring AOM-susceptibility modifier loci. The first method used three-SNP (single nucleotide polymorphism) windows to infer haplotype structure among the strains (McClurg et al. 2006, 2007). The inferred haplotypes were used to perform association analysis. The second method was a tree-based method, which makes tree hierarchies derived from SNPs with a compatible interval based on 7 million SNPs (Pan et al. 2009). This approach performs association analysis using all possible groupings based on a tree hierarchy.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All data are present in the manuscript and is freely available. Mouse SNPs are in public databases.
Results
Extensive interstrain variation in susceptibility to AOM-induced CRC
Treatment of a genetically diverse population of mouse inbred strains with AOM results in a continuous distribution in tumor susceptibility. Although tumor penetrance (percent of mice with one or more tumors; Figure 1A) and average multiplicity (average number of tumors per tumor-bearing mouse; Figure 1B) were highly correlated (r2 = 0.88), mean tumor size was less correlated with the other measures, suggesting independent genetic control. Tumor size ranged from 1.5 to 5.25 mm in mean diameter (Figure 1C). Of the 40 strains tested, 10 were completely resistant to AOM, while 18 strains exhibited a tumor penetrance >25%. There was no correlation between genealogy and tumor incidence; the most sensitive strains, including KK/H1J, C57 L/J, A/J, and MOLF/EiJ, were derived from diverse genealogies (Bogue and Grubb 2004). Nonetheless, the wild-derived strains exhibited the most similarity in tumor susceptibility, with six of the seven wild-derived strains exhibiting resistance to AOM. The only exception was the Mus musculus molossinus-derived strain MOLF/EiJ, which was among the most susceptible. However, the other two M. m. molossinus-derived strains, JF1/MsJ and MSM/MsJ, were resistant to AOM. The C57-related strains showed varying degrees of sensitivity to AOM with C57BL/6J being the least sensitive and C57L/J being the most sensitive.
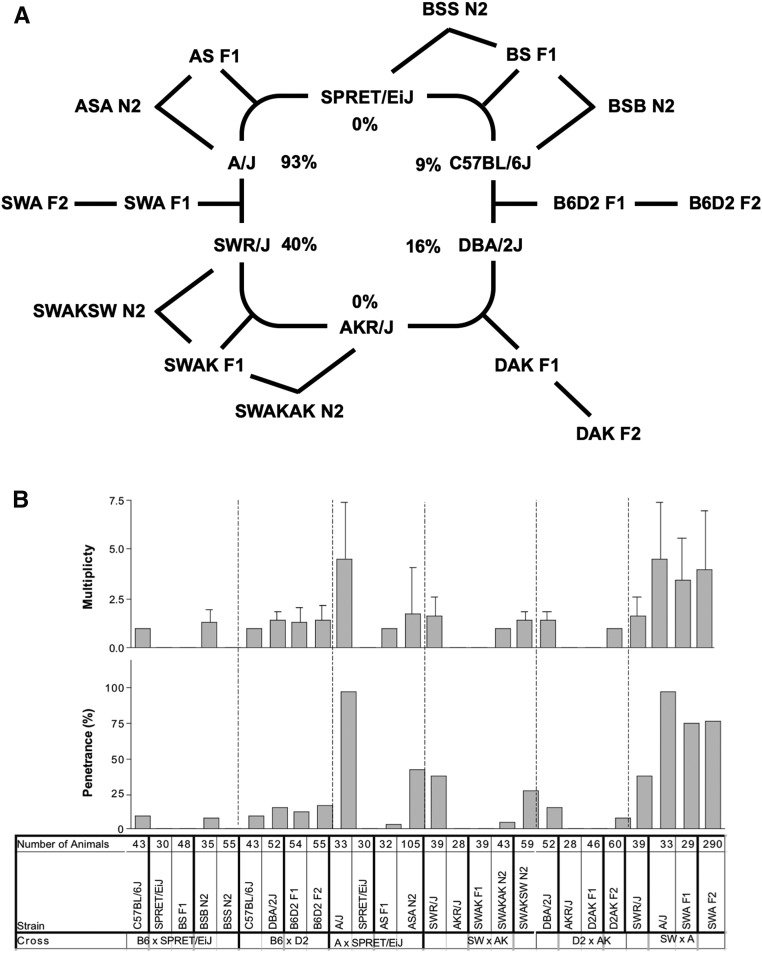
Figure 1.
Variable response of mouse strains to azoxymethane. (A) Penetrance of colon tumors. (B) Average number of colon tumors in those mice with one or more tumor. (C) Average diameter of colon tumors in tumor bearing mice. Number of mice used from each strain and distributed by sex is noted. (B) and (C) are mean +/− SE.
The distribution of tumors along the length of the colon demonstrated that the majority of tumors develop in the distal half (Figure 2A). MOLF/EiJ mice had the greatest proportion of tumors in the proximal half of the colon. Although tumor incidence was slightly higher in females than in males, tumor penetrance between sexes was highly correlated (r2 = 0.83; Figure 2B). Despite the high correlation, several strains did show differences in tumor penetrance between males and females, but these did not result in statistical differences in any other tumor characteristic. The greatest difference was observed for the P/J strain, where female mice had a threefold higher tumor penetrance than males. Sex-specific penetrance with C57BL/6J and LG/J were most pronounced with females and males, respectively, having almost 20% penetrance, while the alternative sex within the respective strains developed no tumors.
Figure 2.
Strains have characteristic responses to azoxymethane. (A) Location of tumors throughout the colon in those strains with at least one tumor-bearing mouse. (B) Penetrance of colon tumors separated by sex in tumor bearing strains.
Resistance to CRC development shows genome-wide dominance and polygenic inheritance
Genetic crosses between strains with similar and varying levels of susceptibility were performed to further investigate the genetic architecture of susceptibility to AOM-induced carcinogenesis (Figure 3A). Tumor penetrance in F1 hybrids between susceptible and resistant strains revealed strong, genome-wide dominance for resistance to AOM-induced carcinogenesis; crosses between susceptible and resistant strains including C57BL/6J and SPRET/EiJ or SWR/J and AKR/J resulted in resistant F1 hybrids and resistant progeny in backcrosses to the resistant parent (Figure 3B). F2 intercross offspring or N2 backcross offspring from F1 mice backcrossed to susceptible parental strains led to tumor incidences that were intermediate between the parental strains. Intermediate susceptibility was also observed in crosses between susceptible strains; F1 and F2 offspring between DBA/J and C57BL/6J, or between SWR/J and A/J, resulted in tumor penetrance and multiplicity intermediate to that of the parental strains. As such, tumor incidence did not segregate in a Mendelian fashion in any intercrosses or backcrosses, suggesting involvement of multiple alleles.
Figure 3.
Resistance to colon carcinogenesis is dominant. (A) Crosses analyzed between strains with penetrance of inbred strains used in the crosses. (B) Penetrance and multiplicity (average tumor number) in each cross showing the number of mice used for the analysis.
Multiple alleles contribute to AOM-induced carcinogenesis susceptibility
Previous mapping studies have reported 24 modifier alleles that contribute to the susceptibility of DMH or AOM-induced CRC (Moen et al. 1992, 1996; Jacoby et al. 1994; van Wezel et al. 1999; Angel et al. 2000; Ruivenkamp et al. 2003; Meunier et al. 2010, 2011, 2013; Eversley et al. 2012; Liu et al. 2012). Of the 24 modifiers, Colon cancer susceptibility 2 (Ccs2) overlaps with Susceptibility to colon cancer 7 (Scc7) on Chromosome (Chr) 3, while the remaining modifiers are distributed across 13 different chromosomes (Angel and DiGiovanni 2018). To extend the number, and to narrow the location of CRC modifiers, haplotype association mapping (HAM) (McClurg et al. 2006, 2007) and tree-based association mapping (Pan et al. 2009) were performed using dense SNP maps and AOM susceptibility data from the inbred strain panel.
HAM treats each inbred strain as one sample, and uses contiguous three-SNP windows to infer haplotype structure across strains. Subsequently, the inferred haplotypes are used to perform association analysis. Within the 40 inbred strains, there are several wild-derived strains. Because the genomes of wild-derived strains are substantially different from laboratory strains, they tend to have distinct haplotypes across much of their genome, leading to noise in HAM. Additionally, genotype data for several laboratory strains are not available. After removing the wild-derived and laboratory strains lacking dense genotype maps, 28 inbred strains were available for HAM.
Since tumor penetrance of <10% accounts for 52.5% of the 28 strains, the data were transformed into a categorical variable. Tumor multiplicity and tumor size was integrated by multiplying mean tumor multiplicity with mean tumor size for each strain to generate a tumor load measurement. HAM was then performed for categorical penetrance, original penetrance, mean tumor size, mean tumor multiplicity, and tumor load (Figure 4A; Table 1). HAM typically leads to relatively high false-positive rates without informative P-values due to limited sample sizes and population substructures within laboratory strains (McClurg et al. 2006). Therefore, the relative highest value would be more informative compared to the absolute association score. Additionally, local regions tend to have similar association values because of linkage disequilibrium. The 25 1-Mb intervals with the largest association scores were plotted as candidate intervals harboring AOM susceptibility modifiers to compare their locations with previously reported CRC susceptibility loci (Figure 4B). HAM results detect loci overlapping with those previously detected, including loci on Chr 4 (Scc11), 7 (Scc12), and 11 (Scc6), indicating that the underlying genes could be the same. There is also a positive signal on Chr 4 at ∼145 Mb, which coincides with the location of Kras—a gene mutated in about half of human CRCs.
Figure 4.
Haplotype association mapping of loci influencing colon carcinogenesis. (A) Distribution of statistical associations (y-axis) between colon tumor phenotypes and genomic region (x-axis). (B) Map of colon tumor modifier locations compared to previously mapped modifiers (green bars). Modifiers for categorical penetrance (red), original penetrance (blue) mean tumor size (purple), mean tumor multiplicity (yellow), and tumor load (green) are marked by squares.
Table 1. Locations of the top 25 log.p scores for each colon carcinogenesis trait and the previous loci that have been mapped to overlapping intervals.
| Categorical Penetrance | Original Penetrance | ||||||
|---|---|---|---|---|---|---|---|
| Chromosome | Position (Mb) | log.p | Locus | Chromosome | Position (Mb) | log.p | Locus |
| 1 | 9926281 | 2.9 | 1 | 9070897 | 2.5 | ||
| 1 | 56753998 | 2.8 | Scc20 | 2 | 109029948 | 2.9 | Scc10 |
| 1 | 73441081 | 2.8 | 3 | 148884697 | 2.8 | Scc7 | |
| 1 | 168920258 | 2.9 | 4 | 12234839 | 2.7 | ||
| 2 | 62446341 | 2.8 | 4 | 123308576 | 3.0 | Scc11 | |
| 2 | 63131035 | 3.0 | Scc2 | 6 | 145773921 | 3.4 | Scc25a |
| 2 | 68102376 | 2.7 | 7 | 112294399 | 2.5 | ||
| 2 | 151395251 | 2.8 | Scc10 | 7 | 127454250 | 2.9 | Scc12 |
| 4 | 35895577 | 2.8 | 8 | 82818143 | 2.7 | ||
| 4 | 37474707 | 2.7 | 8 | 119408251 | 2.5 | ||
| 4 | 42909029 | 3.1 | Scc22a | 8 | 119967314 | 2.7 | |
| 4 | 4344933 | 2.8 | 9 | 57763899 | 2.7 | Ccs5 | |
| 4 | 44056939 | 2.9 | 10 | 117526565 | 2.5 | Scc9 | |
| 4 | 48108791 | 2.9 | 11 | 16803015 | 2.9 | Scc6 | |
| 4 | 152525707 | 2.9 | 11 | 22114857 | 2.6 | ||
| 5 | 70446395 | 2.7 | 11 | 89399589 | 2.6 | ||
| 5 | 114567323 | 2.7 | 14 | 5986996 | 2.7 | ||
| 5 | 115137232 | 3.6 | Scc2a | 14 | 119802709 | 2.9 | |
| 5 | 115818557 | 3.2 | 14 | 120417186 | 3.0 | Scc26a | |
| 5 | 124021828 | 2.7 | 15 | 68242489 | 2.8 | ||
| 6 | 145493742 | 2.9 | 15 | 96028694 | 2.7 | ||
| 7 | 139840681 | 2.8 | Scc12 | 18 | 440044 | 2.6 | |
| 9 | 115593372 | 3.0 | Scc24a | X | 61860172 | 2.9 | |
| 10 | 27493984 | 2.8 | Scc14 | X | 63148384 | 2.9 | |
| 11 | 15664968 | 2.7 | Scc6 | X | 64149116 | 2.9 | |
| Mean Tumor Size | Mean Tumor Multiplicity | ||||||
|---|---|---|---|---|---|---|---|
| Chromosome | Position (Mb) | log.p | Locus | Chromosome | Position (Mb) | log.p | Locus |
| 1 | 51206257 | 2.4 | Scc20 | 1 | 90708743 | 2.7 | |
| 2 | 26233654 | 2.3 | 1 | 177991709 | 2.4 | Scc3 | |
| 3 | 73362679 | 2.4 | 1 | 182638381 | 2.4 | ||
| 3 | 74123762 | 2.8 | 2 | 109029948 | 2.6 | Scc10 | |
| 4 | 109270585 | 2.4 | 3 | 135933677 | 2.4 | Ccs3 | |
| 4 | 120383743 | 3.3 | Scc11 | 3 | 148884697 | 2.8 | Scc7 |
| 4 | 123397963 | 2.4 | 4 | 12234839 | 2.4 | ||
| 4 | 155013209 | 2.7 | 4 | 12331322 | 2.7 | Scc11 | |
| 5 | 64846314 | 2.3 | 6 | 145767248 | 3.0 | Scc25a | |
| 5 | 87847984 | 2.4 | 7 | 127454250 | 2.6 | Scc12 | |
| 5 | 88540008 | 2.4 | 8 | 82818143 | 2.6 | ||
| 6 | 143254941 | 2.3 | 8 | 118970189 | 2.4 | ||
| 7 | 76027782 | 2.4 | 8 | 119967314 | 2.7 | ||
| 7 | 131560996 | 2.4 | 9 | 53996096 | 2.4 | ||
| 7 | 132839711 | 2.7 | Scc12 | 9 | 57763899 | 3.0 | Ccs5 |
| 8 | 78611026 | 2.8 | 10 | 117526565 | 2.8 | Scc9 | |
| 8 | 93312083 | 2.8 | 11 | 16803015 | 2.5 | Scc6 | |
| 9 | 20458329 | 2.4 | 11 | 22114857 | 2.6 | ||
| 11 | 116815005 | 2.6 | Scc16 | 11 | 89356815 | 2.4 | |
| 13 | 81611768 | 2.4 | 14 | 119802709 | 2.4 | ||
| 15 | 10493883 | 2.5 | 14 | 120417186 | 2.5 | ||
| 15 | 13278872 | 2.4 | 15 | 14502057 | 2.3 | ||
| 16 | 55423042 | 2.4 | 15 | 68242633 | 2.4 | ||
| 19 | 21499147 | 2.4 | 15 | 96028879 | 2.3 | ||
| 19 | 24915159 | 2.3 | 18 | 2510135 | 2.5 | Scc5 | |
| Tumor load | |||||||
| Chromosome | Position (Mb) | log.p | Locus | ||||
| 1 | 19882037 | 2.1 | |||||
| 1 | 191292260 | 2.2 | Scc3 | ||||
| 3 | 79429335 | 2.2 | |||||
| 4 | 124318989 | 2.2 | Scc11 | ||||
| 4 | 152525707 | 2.7 | |||||
| 5 | 114567323 | 2.5 | |||||
| 5 | 115227317 | 2.6 | |||||
| 7 | 125786945 | 2.3 | Scc12 | ||||
| 7 | 126466181 | 2.1 | |||||
| 8 | 28733060 | 2.3 | Scc17 | ||||
| 8 | 76851411 | 2.4 | |||||
| 9 | 115235730 | 2.6 | |||||
| 11 | 19865602 | 2.2 | Scc6 | ||||
| 11 | 20853304 | 2.1 | |||||
| 11 | 21983462 | 2.2 | |||||
| 12 | 74808910 | 2.2 | |||||
| 12 | 75706999 | 2.3 | Ccs1 | ||||
| 13 | 59969925 | 2.6 | |||||
| 15 | 11729102 | 2.7 | |||||
| 15 | 20038911 | 2.2 | |||||
| 17 | 43879545 | 2.2 | |||||
| 17 | 44592447 | 3.0 | Scc27a | ||||
| 17 | 86376062 | 2.2 | Scc4 | ||||
| 18 | 31370952 | 2.4 | Scc5 | ||||
| 18 | 32617091 | 2.4 | |||||
New loci detected here.
A tree-based association mapping approach was also performed using all possible strain groupings based upon an underlying relationship or tree structure (Pan et al. 2009). Consequently, tree-based methods have the potential to identify additional associations that are not detected by HAM. While HAM uses weighted bootstraps to simulate background distributions, the tree-based method implies the F-distribution as the background distribution. As such, tree-based methods would lead to more false positives compared with HAM, and, therefore, are best performed on local regions where pre-existing data suggests modifiers reside.
Tree-based analysis using original penetrance and mean tumor multiplicity on Chr 4 and 6 confirmed the locations determined by HAM (Figure 5A). When the tree structure was analyzed in detail for the association peak on Chr 4, the strain grouping revealed that most susceptible strains, such as KK/H1J and A/J, cluster on one branch, while the other strains are dispersed on several branches (Figure 5B). This result indicates that susceptible strains might share the same susceptibility gene variant. A similar trend from the tree structure is also observed for the Chr 6 locus (Figure 5C).
Figure 5.
Tree-based mapping of loci influencing colon carcinogenesis. (A) Genome-wide distribution of statistical associations. (B) Strains distributed on tree structure for modifier on Chr 4. (C) Strains distributed on tree structure for Chr 6.
In addition to detecting most previously mapped CRC modifiers in mice, six new loci were detected that reached statistical significance (log.P ≥ 3.0; Table 1). These include Scc22 (Chr 4, 42909029Mb), Scc23 (Chr 5, 115137232Mb), and Scc24 (Chr 9, 115593372) detected for categorical penetrance; Scc25 (Chr 6, 145773921Mb) and Scc26 (Chr 14, 120417186Mb) for original penetrance; Scc25 for mean tumor multiplicity; and Scc27 (Chr 17, 44592447Mb) for tumor load. Those loci not given names are suggestive based on their P-values.
Discussion
Epidemiological studies suggest that, although environmental factors are important contributors to human cancer development, genetic susceptibility still plays an important role in nonfamilial or sporadic CRC (Perera 1996; Tomlinson et al. 2007, 2008; Zanke et al. 2007; COGENT Study et al. 2008; Tenesa et al. 2008; Tenesa and Dunlop 2009; Migliore et al. 2011; Theodoratou et al. 2012; Carethers and Jung 2015; Schumacher et al. 2015; Montazeri et al. 2016; Tanikawa et al. 2018; Bien et al. 2019; Lu et al. 2019). The interstrain variation in susceptibility to AOM-induced carcinogenesis supports an important role for genetic modifiers in nonfamilial CRC, as revealed by analysis of AOM susceptibility across genetically heterogeneous mouse strains in a common environment showing substantial genetic variation in CRC susceptibility. Inclusion of such a large number of strains, equivalent to a genetically diverse human population, has provided a wide range of susceptibilities that will be useful for identifying cancer modifier genes, and for supporting selection of strains for additional molecular analysis (Yang et al. 2019a; Zhou and You 2019).
Analysis of genetic crosses between resistant and susceptible strains suggests the involvement of multiple genes that contribute to AOM susceptibility (Ruivenkamp et al. 2003; Meunier et al. 2010, 2011, 2013; Eversley et al. 2012; Liu et al. 2012; Angel and DiGiovanni 2018), consistent with GWAS in humans (Tomlinson et al. 2007, 2008; Zanke et al. 2007; COGENT Study et al. 2008; Tenesa et al. 2008; Tenesa and Dunlop 2009; Theodoratou et al. 2012; Schumacher et al. 2015; Montazeri et al. 2016; Tanikawa et al. 2018; Bien et al. 2019; Lu et al. 2019). Crosses between susceptible and resistant strains manifest a resistant phenotype in both F1 mice and N2 progeny generated by backcrossing F1 mice to their resistant parental strain. F2 and N2 progeny generated by backcrossing F1 mice to their susceptible parental strain frequently exhibited susceptibilities intermediate to that of their respective parental strains. Consequently, alleles conferring resistance are not as strong, or are not present in high numbers in susceptible strains since F1 and F2 progeny of crosses between susceptible strains exhibited intermediate susceptibility. These results indicate that complementary resistance alleles are usually not present in susceptible strains. These results also indicate that, on a genome-wide scale, cancer resistance is dominant, and that susceptible individuals are likely lacking sufficient numbers of resistance alleles to reduce cancer incidence rather than having specific cancer susceptibility alleles. This observation may explain why most individuals, even in environments with elevated cancer rates, never develop CRC, and has important implications on how genome-wide studies for cancer susceptibility are analyzed in humans. Based on the AOM-susceptibility results, GWAS in humans are likely to have increased power when analyzed for absence of multiple resistance alleles rather than for differential allele frequencies in populations of cancer cases vs. controls as is commonly done. Similarly, the identification of patients with heightened risk for CRC should have greater distinguishing power if individuals are categorized based upon the number of resistance alleles they harbor, rather than whether they have specific susceptibility alleles.
Based on overlapping localizations, all previous CRC modifier alleles in mice were detected in the current association study except for six: Scc1, Scc8, Scc13, Scc15, Scc18, and Scc21 (Table 1), providing confirmation of the existence of most previously detected alleles. Several previous mapping studies used CcS/Dem recombinant congenic (RC) strains treated with DMH to identify colon cancer susceptibility loci (Moen et al. 1992, 1996; van Wezel et al. 1999). The CcS/Dem RC strains consist of a background strain, BALB/cHeA (DMH resistant) and a donor strain, STS/A (DMH susceptible) that comprise 87.5 and 12.5% of the RC genomes, respectively (Moen et al. 1991). Three of the CcS strains (CcS 7, 11, and 19) were used in the present study, and their relative tumor penetrance matched that observed in response to DMH, with CcS 19 being the most susceptible (Moen et al. 1991). DMH treatment of CcS RC strains or crosses of CcS 19 with BALB/c was used to identify tumor susceptibility loci Scc1, Scc2, and Scc6-9 (Moen et al. 1992, 1996; van Wezel et al. 1999). Three additional loci (Scc3-5) were identified in (BALB/c x CcS 19) F2 mice treated with a combination of DMH and N-ethyl-N-nitrosourea (van Wezel et al. 1996). Analyses of (BALB/c x CcS 19) F2 mice treated with AOM identified Scc11-15 (Ruivenkamp et al. 2003). Additional CRC susceptibility loci (Ccs1-Ccs3, Ccs5) have been identified in crosses of sensitive and resistant laboratory mouse strains treated with DMH or AOM (Jacoby et al. 1994; Angel et al. 2000; Meunier et al. 2010, 2011, 2013) and in interspecific backcrosses of resistant SPRET/EiJ with sensitive A/J mice treated with AOM (Scc16-21) (Eversley et al. 2012). A number of GWAS have been conducted in humans (Tomlinson et al. 2007, 2008; Zanke et al. 2007; COGENT Study et al. 2008; Tenesa et al. 2008; Schumacher et al. 2015; Tanikawa et al. 2018; Bien et al. 2019; Lu et al. 2019), which revealed several common variants that function as low-penetrance susceptibility alleles. The results of the current strain characterization provide a foundation for extending the catalog of CRC modifier alleles that can be used to evaluate the spectrum of resistances among individuals. Future identification of the underlying genes responsible for the Scc loci should reveal the relationship between mouse and human cancer susceptibility, and how genetic modifiers influence susceptibility.
Acknowledgments
We thank the many members of the Threadgill laboratory who assisted with mouse husbandry and necropsies throughout the project, and for suggestions and comments on the manuscript. This work was supported by National Cancer Institute grants (CA079869, CA105417, and CA106991 to D.W.T.) and a National Institute of Environmental Health Sciences fellowship (ES012354 to A.C.B.). Author contributions: D.W.T. and R.S.P. designed the study concept and experiments; R.W.E. and P.D. provided mouse strains and advised on study design; A.C.B., R.S.P., D.K., and D.W.T performed the study; Y.X., L.Y., L.M., F.P-M.deV, and D.W.T. performed data analysis; A.C.B. and D.W.T. wrote the manuscript; J.M.A. edited the manuscript, and all authors read the manuscript.
Footnotes
Communicating editor: S. Sharan
Literature Cited
- Angel J. M., and DiGiovanni J., 2018. Genetic determinants of cancer susceptibility, pp. 330–360 in Comprehensive toxicology, Ed. 3, edited by McQueen C. A., Elsevier Science, Oxford: 10.1016/B978-0-12-801238-3.65251-0 [DOI] [Google Scholar]
- Angel J. M., Popova N., Lanko N., Turusov V. S., and DiGiovanni J., 2000. A locus that influences susceptibility to 1, 2-dimethylhydrazine-induced colon tumors maps to the distal end of mouse chromosome 3. Mol. Carcinog. 27: 47–54. [DOI] [PubMed] [Google Scholar]
- Bi W., Liu H., Shen J., Zhang L. H., Li P. et al. , 2017. Chemopreventive effects of Ku-jin tea against AOM-induced precancerous colorectal lesions in rats and metabolomic analysis. Sci. Rep. 7: 15893 10.1038/s41598-017-16237-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien S. A., Su Y. R., Conti D. V., Harrison T. A., Qu C. et al. , 2019. Genetic variant predictors of gene expression provide new insight into risk of colorectal cancer. Hum. Genet. 138: 307–326 (erratum: Hum. Genet. 138: 789–791). 10.1007/s00439-019-01989-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissahoyo A., Pearsall R. S., Hanlon K., Amann V., Hicks D. et al. , 2005. Azoxymethane is a genetic background-dependent colorectal tumor initiator and promoter in mice: effects of dose, route, and diet. Toxicol. Sci. 88: 340–345. 10.1093/toxsci/kfi313 [DOI] [PubMed] [Google Scholar]
- Bogue M. A., and Grubb S. C., 2004. The mouse phenome project. Genetica 122: 71–74. 10.1007/s10709-004-1438-4 [DOI] [PubMed] [Google Scholar]
- Burt R. W., Bishop D. T., Cannon-Albright L., Samowitz W. S., Lee R. L. et al. , 1992. Population genetics of colonic cancer. Cancer 70: 1719–1722. [DOI] [PubMed] [Google Scholar]
- Cancer Facts and Figures, 2019 Leading sites of new cancer cases and deaths—2019 estimates. American Cancer Society. Available at: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2019/leading-sites-of-new-cancer-cases-and-deaths-2019-estimates.pdf. Accessed: September 1, 2019.
- Carethers J. M., and Jung B. H., 2015. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 149: 1177–1190 e3. 10.1053/j.gastro.2015.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., and Huang X. F., 2009. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol. Ther. 8: 1313–1317. 10.4161/cbt.8.14.8983 [DOI] [PubMed] [Google Scholar]
- COGENT Study, Houlston R. S., Webb E., Broderick P., Pittman A. M., Di Bernardo M. C. et al. , 2008. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 40: 1426–1435. 10.1038/ng.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckrey H., Preussmann R., Matzkies F., and Ivankovic S., 1967. Selektive Erzeugung von Darmkrebs bei Ratten durch 1,2-Dimethylhydrozin. Naturwissenschaften 54: 285–286. 10.1007/BF00620890 [DOI] [PubMed] [Google Scholar]
- Eversley C. D., Yuying X., Pearsall R. S., and Threadgill D. W., 2012. Mapping six new susceptibility to colon cancer (Scc) loci using a mouse interspecific backcross. G3 (Bethesda) 2: 1577–1584. 10.1534/g3.112.002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Watkins P. B., Su S., Ross P. K., Harbourt D. E. et al. , 2009. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 19: 1507–1515. 10.1101/gr.090241.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter C. M., Chang-Claude J., Slattery M. L., Pflugeisen B. M., Lin Y. et al. , 2012. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 72: 2036–2044. 10.1158/0008-5472.CAN-11-4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby R. F., Hohman C., Marshall D. J., Frick T. J., Schlack S. et al. , 1994. Genetic analysis of colon cancer susceptibility in mice. Genomics 22: 381–387. 10.1006/geno.1994.1399 [DOI] [PubMed] [Google Scholar]
- Kaiser S., Park Y. K., Franklin J. L., Halberg R. B., Yu M. et al. , 2007. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8: R131 10.1186/gb-2007-8-7-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Lu Y., Liu H., Wen W., Jia D. et al. , 2012. Genome-wide association and fine mapping of genetic loci predisposing to colon carcinogenesis in mice. Mol. Cancer Res. 10: 66–74. 10.1158/1541-7786.MCR-10-0540 [DOI] [PubMed] [Google Scholar]
- Liu J., Zheng B., Li Y., Yuan Y., and Xing C., 2019. Genetic polymorphisms of DNA repair pathways in sporadic colorectal carcinogenesis. J. Cancer 10: 1417–1433. 10.7150/jca.28406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Kweon S. S., Tanikawa C., Jia W. H., Xiang Y. B. et al. , 2019. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology 156: 1455–1466. 10.1053/j.gastro.2018.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S. K., Golla S., Golla J. P., Tanaka N., Cai Y. et al. , 2015. St. John’s wort attenuates colorectal carcinogenesis in mice through suppression of inflammatory signaling. Cancer Prev. Res. (Phila.) 8: 786–795. 10.1158/1940-6207.CAPR-14-0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClurg P., Pletcher M. T., Wiltshire T., and Su A. I., 2006. Comparative analysis of haplotype association mapping algorithms. BMC Bioinformatics 7: 61 10.1186/1471-2105-7-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClurg P., Janes J., Wu C., Delano D. L., Walker J. R. et al. , 2007. Genomewide association analysis in diverse inbred mice: power and population structure. Genetics 176: 675–683. 10.1534/genetics.106.066241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Md Nasir N. L., Kamsani N. E., Mohtarrudin N., Othman F., Md Tohid S. F. et al. , 2017. Anticarcinogenic activity of Muntingia calabura leaves methanol extract against the azoxymethane-induced colon cancer in rats involved modulation of the colonic antioxidant system partly by flavonoids. Pharm. Biol. 55: 2102–2109. 10.1080/13880209.2017.1371769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier C., Cai J., Fortin A., Kwan T., Marquis J. F. et al. , 2010. Characterization of a major colon cancer susceptibility locus (Ccs3) on mouse chromosome 3. Oncogene 29: 647–661. 10.1038/onc.2009.369 [DOI] [PubMed] [Google Scholar]
- Meunier C., Kwan T., Turbide C., Beauchemin N., and Gros P., 2011. Genetic control of susceptibility to carcinogen-induced colorectal cancer in mice: the Ccs3 and Ccs5 loci regulate different aspects of tumorigenesis. Cell Cycle 10: 1739–1749. 10.4161/cc.10.11.15817 [DOI] [PubMed] [Google Scholar]
- Meunier C., Van Der Kraak L., Turbide C., Groulx N., Labouba I. et al. , 2013. Positional mapping and candidate gene analysis of the mouse Ccs3 locus that regulates differential susceptibility to carcinogen-induced colorectal cancer. PLoS One 8: e58733 10.1371/journal.pone.0058733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore L., Migheli F., Spisni R., and Coppede F., 2011. Genetics, cytogenetics, and epigenetics of colorectal cancer. J. Biomed. Biotechnol. 2011: 792362 10.1155/2011/792362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen C. J., van der Valk M. A., Snoek M., van Zutphen B. F., von Deimling O. et al. , 1991. The recombinant congenic strains–a novel genetic tool applied to the study of colon tumor development in the mouse. Mamm. Genome 1: 217–227. 10.1007/BF00352328 [DOI] [PubMed] [Google Scholar]
- Moen C. J., Snoek M., Hart A. A., and Demant P., 1992. Scc-1, a novel colon cancer susceptibility gene in the mouse: linkage to CD44 (Ly-24, Pgp-1) on chromosome 2. Oncogene 7: 563–566. [PubMed] [Google Scholar]
- Moen C. J., Groot P. C., Hart A. A., Snoek M., and Demant P., 1996. Fine mapping of colon tumor susceptibility (Scc) genes in the mouse, different from the genes known to be somatically mutated in colon cancer. Proc. Natl. Acad. Sci. USA 93: 1082–1086. 10.1073/pnas.93.3.1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montazeri Z., Theodoratou E., Nyiraneza C., Timofeeva M., Chen W. et al. , 2016. Systematic meta-analyses and field synopsis of genetic association studies in colorectal adenomas. Int. J. Epidemiol. 45: 186–205. 10.1093/ije/dyv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser A. R., Pitot H. C., and Dove W. F., 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247: 322–324. 10.1126/science.2296722 [DOI] [PubMed] [Google Scholar]
- Nambiar P. R., Girnun G., Lillo N. A., Guda K., Whiteley H. E. et al. , 2003. Preliminary analysis of azoxymethane induced colon tumors in inbred mice commonly used as transgenic/knockout progenitors. Int. J. Oncol. 22: 145–150. [PubMed] [Google Scholar]
- Odun-Ayo F., Mellem J., Naicker T., and Reddy L., 2015. Chemoprevention of azoxymethane-induced colonic carcinogenesis in balb/c mice using a modified pectin alginate probiotic. Anticancer Res. 35: 4765–4775. [PubMed] [Google Scholar]
- Pan F., McMillan L., Pardo-Manuel De Villena F., Threadgill D., and Wang W., 2009. TreeQA: quantitative genome wide association mapping using local perfect phylogeny trees. Pac. Symp. Biocomput. 2009: 415–426. [PMC free article] [PubMed] [Google Scholar]
- Pedro D. F., Ramos A. A., Lima C. F., Baltazar F., and Pereira-Wilson C., 2016. Colon cancer chemoprevention by sage tea drinking: decreased DNA damage and cell proliferation. Phytother. Res. 30: 298–305. 10.1002/ptr.5531 [DOI] [PubMed] [Google Scholar]
- Perera F. P., 1996. Molecular epidemiology: insights into cancer susceptibility, risk assessment, and prevention. J. Natl. Cancer Inst. 88: 496–509. 10.1093/jnci/88.8.496 [DOI] [PubMed] [Google Scholar]
- Piazzi G., Prossomariti A., Baldassarre M., Montagna C., Vitaglione P. et al. , 2019. A Mediterranean diet mix has chemopreventive effects in a murine model of colorectal cancer modulating apoptosis and the gut microbiota. Front. Oncol. 9: 140 10.3389/fonc.2019.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B. S., 2004. Studies with the azoxymethane-rat preclinical model for assessing colon tumor development and chemoprevention. Environ. Mol. Mutagen. 44: 26–35. 10.1002/em.20026 [DOI] [PubMed] [Google Scholar]
- Ruivenkamp C. A., Csikos T., Klous A. M., van Wezel T., and Demant P., 2003. Five new mouse susceptibility to colon cancer loci, Scc11-Scc15. Oncogene 22: 7258–7260. 10.1038/sj.onc.1207096 [DOI] [PubMed] [Google Scholar]
- Schumacher F. R., Schmit S. L., Jiao S., Edlund C. K., Wang H. et al. , 2015. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat. Commun. 6: 7138 (erratum: Nat. Commun. 6: 8739). 10.1038/ncomms8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzui M., Ushijima T., Dashwood R. H., Yoshimi N., Sugimura T. et al. , 1999. Frequent mutations of the rat beta-catenin gene in colon cancers induced by methylazoxymethanol acetate plus 1-hydroxyanthraquinone. Mol. Carcinog. 24: 232–237. [DOI] [PubMed] [Google Scholar]
- Takahashi M., and Wakabayashi K., 2004. Gene mutations and altered gene expression in azoxymethane-induced colon carcinogenesis in rodents. Cancer Sci. 95: 475–480. 10.1111/j.1349-7006.2004.tb03235.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Hosono K., Endo H., and Nakajima A., 2013. Colon epithelial proliferation and carcinogenesis in diet-induced obesity. J. Gastroenterol. Hepatol. 28: 41–47. 10.1111/jgh.12240 [DOI] [PubMed] [Google Scholar]
- Tanikawa C., Kamatani Y., Takahashi A., Momozawa Y., Leveque K. et al. , 2018. GWAS identifies two novel colorectal cancer loci at 16q24.1 and 20q13.12. Carcinogenesis 39: 652–660. 10.1093/carcin/bgy026 [DOI] [PubMed] [Google Scholar]
- Tenesa A., and Dunlop M. G., 2009. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat. Rev. Genet. 10: 353–358. 10.1038/nrg2574 [DOI] [PubMed] [Google Scholar]
- Tenesa A., Farrington S. M., Prendergast J. G., Porteous M. E., Walker M. et al. , 2008. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 40: 631–637. 10.1038/ng.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoratou E., Montazeri Z., Hawken S., Allum G. C., Gong J. et al. , 2012. Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J. Natl. Cancer Inst. 104: 1433–1457. 10.1093/jnci/djs369 [DOI] [PubMed] [Google Scholar]
- Tomlinson I., Webb E., Carvajal-Carmona L., Broderick P., Kemp Z. et al. , 2007. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 39: 984–988. 10.1038/ng2085 [DOI] [PubMed] [Google Scholar]
- Tomlinson I. P., Webb E., Carvajal-Carmona L., Broderick P., Howarth K. et al. , 2008. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 40: 623–630. 10.1038/ng.111 [DOI] [PubMed] [Google Scholar]
- Tulchin N., Ornstein L., Guillem J., O’Toole K., Lambert M. E. et al. , 1988. Distribution of the c-myc oncoprotein in normal and neoplastic tissues of the rat colon. Oncogene 3: 697–701. [PubMed] [Google Scholar]
- van Wezel T., Stassen A. P., Moen C. J., Hart A. A., van der Valk M. A. et al. , 1996. Gene interaction and single gene effects in colon tumour susceptibility in mice. Nat. Genet. 14: 468–470. 10.1038/ng1296-468 [DOI] [PubMed] [Google Scholar]
- van Wezel T., Ruivenkamp C. A., Stassen A. P., Moen C. J., and Demant P., 1999. Four new colon cancer susceptibility loci, Scc6 to Scc9 in the mouse. Cancer Res. 59: 4216–4218. [PubMed] [Google Scholar]
- Waly M. I., Al-Rawahi A. S., Al Riyami M., Al-Kindi M. A., Al-Issaei H. K. et al. , 2014. Amelioration of azoxymethane induced-carcinogenesis by reducing oxidative stress in rat colon by natural extracts. BMC Complement. Altern. Med. 14: 60 10.1186/1472-6882-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. S., Papanikolaou A., Sabourin C. L., and Rosenberg D. W., 1998. Altered expression of cyclin D1 and cyclin-dependent kinase 4 in azoxymethane-induced mouse colon tumorigenesis. Carcinogenesis 19: 2001–2006. 10.1093/carcin/19.11.2001 [DOI] [PubMed] [Google Scholar]
- Wu X., Song M., Qiu P., Rakariyatham K., Li F. et al. , 2017. Synergistic chemopreventive effects of nobiletin and atorvastatin on colon carcinogenesis. Carcinogenesis 38: 455–464. 10.1093/carcin/bgx018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bell T. A., Churchill G. A., and Pardo-Manuel de Villena F., 2007. On the subspecific origin of the laboratory mouse. Nat. Genet. 39: 1100–1107. 10.1038/ng2087 [DOI] [PubMed] [Google Scholar]
- Yang C., Wang Y., Xu W., Liu Z., Zhou S. et al. , 2019a Genome-wide association study using diversity outcross mice identified candidate genes of pancreatic cancer. Genomics 111: 1882–1888. 10.1016/j.ygeno.2018.12.011 [DOI] [PubMed] [Google Scholar]
- Yang T., Li X., Montazeri Z., Little J., Farrington S. M. et al. , 2019b Gene-environment interactions and colorectal cancer risk: an umbrella review of systematic reviews and meta-analyses of observational studies. Int. J. Cancer 145: 2315–2329. 10.1002/ijc.32057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanke B. W., Greenwood C. M., Rangrej J., Kustra R., Tenesa A. et al. , 2007. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 39: 989–994. 10.1038/ng2089 [DOI] [PubMed] [Google Scholar]
- Zhou Y., and You M., 2019. Integrative system genetic analysis reveals mRNA-lncRNA network associated with mouse spontaneous lung cancer susceptibility. Oncotarget 10: 339–351. 10.18632/oncotarget.26554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Richardson J. A., Parada L. F., and Graff J. M., 1998. Smad3 mutant mice develop metastatic colorectal cancer. Cell 94: 703–714. 10.1016/S0092-8674(00)81730-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All data are present in the manuscript and is freely available. Mouse SNPs are in public databases.