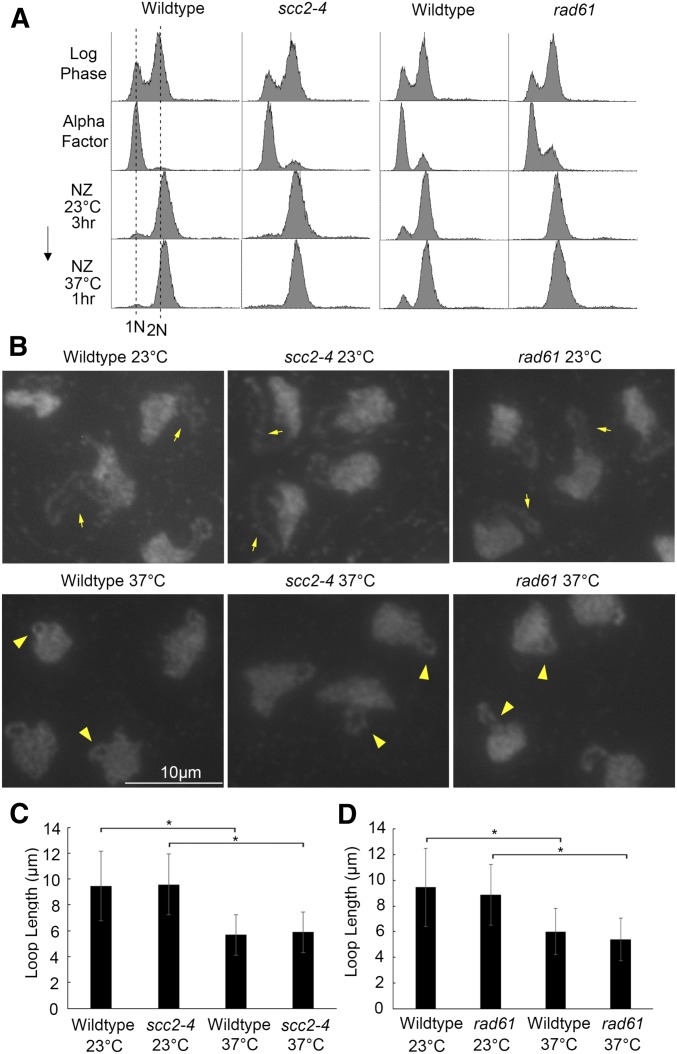
Figure 1.
Cohesin deposition and/or release are not required for hyperthermic-induced rDNA hypercondensation. (A) Flow cytometer data documents DNA content of wild type (YBS1039), scc2-4 mutant cells (YMM511), and rad61 null cells (YBS2037) throughout the experiment. Wild-type and scc2-4 mutant cells were processed separately from wild-type and rad61 null cells. Cells were maintained in nocodazole for 3 hr at 23°, post-alpha factor arrest, followed by an additional 1 hr incubation at 37°. (B) Chromosomal mass and rDNA loop structures detected using DAPI. Yellow arrows indicate long rDNA loops. Yellow arrowheads indicate short rDNA loops. All field of views are shown at equal magnification. (C) Quantification of the loop length of condensed rDNA in wild-type and scc2-4 mutant cells. Data shown was obtained from three biological replicates. For wild type vs. scc2-4 mutant, the results reflect n values of 228 of wild-type cells at 23°, 235 for scc2-4 cells at 23°, 214 for wild-type cells at 37°, and 201 for scc2-4 cells at 37°. (D) Quantification of loop lengths of condensed rDNA in wild-type and rad61 null cells. For wild-type vs. rad61 null mutant, the results reflect n values of 235 for wildtype cells at 23°, 214 for rad61 cells at 23°, 157 for wildtype cells at 37°, and 151 for rad61 cells at 37°. Statistical analysis was performed using a Tukey HSD one way ANOVA. P-value = 0.721 indicates that there is no significant difference between the average loop lengths of wild-type cells (5.67 μm) and scc2-4 mutant cells (5.89 μm) at 37° in (C). P-value = 0.103 indicates there is no significant difference between wild-type cells (6.02 μm) and rad61 null cells (5.40 μm) at 37° in (D). Statistically significant differences (*) are based on P < 0.05.