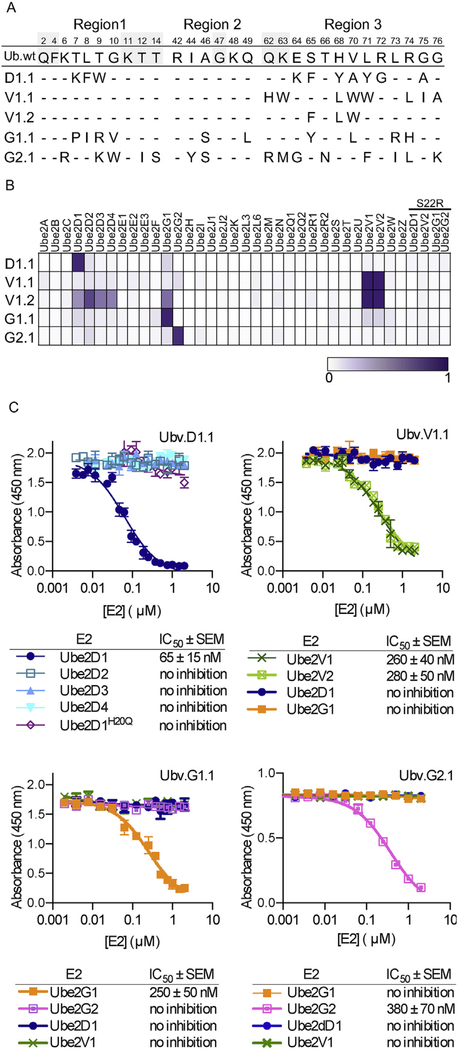
Fig. 2. Selective and tight binding of UbVs to E2 proteins.
(A) Sequence alignment of Ub.wt and UbVs selected for binding to Ube2D1 (D1.1), Ube2V1 (V1.1 and V1.2), Ube2G1 (G1.1), or Ube2G2 (G2.1). The alignment shows only those positions that were diversified in the UbV libraries, and positions that were conserved as the wt sequence are indicated by dashes. (B) The binding specificities of UbVs (y-axis) are shown across the human E2 family (x-axis), as assessed using ELISAs. FLAG-tagged UbV proteins were added to immobilized E2 proteins, and bound UbV proteins were detected by the addition of anti-FLAG-HRP and colorimetric development of TMB peroxidase substrate. The normalized mean value of absorbance at 450 nm is shaded in a purple gradient (white = 0 and purple = 1). “S22R” refers to E2 variants containing an Arg residue in place of Ser22 (Ube2D1 numbering). (C) IC50 values, determined using competition ELISA, for solution-phase E2 proteins inhibiting binding of UbVs to immobilized E2 proteins. Subsaturating concentrations of the indicated UbV proteins were incubated with serial dilutions of E2 proteins as indicated and binding to immobilized cognate E2 protein was detected. The IC50 value was defined as the concentration of solution-phase E2 protein that inhibited 50% of the binding of the UbV protein to the immobilized E2 protein.