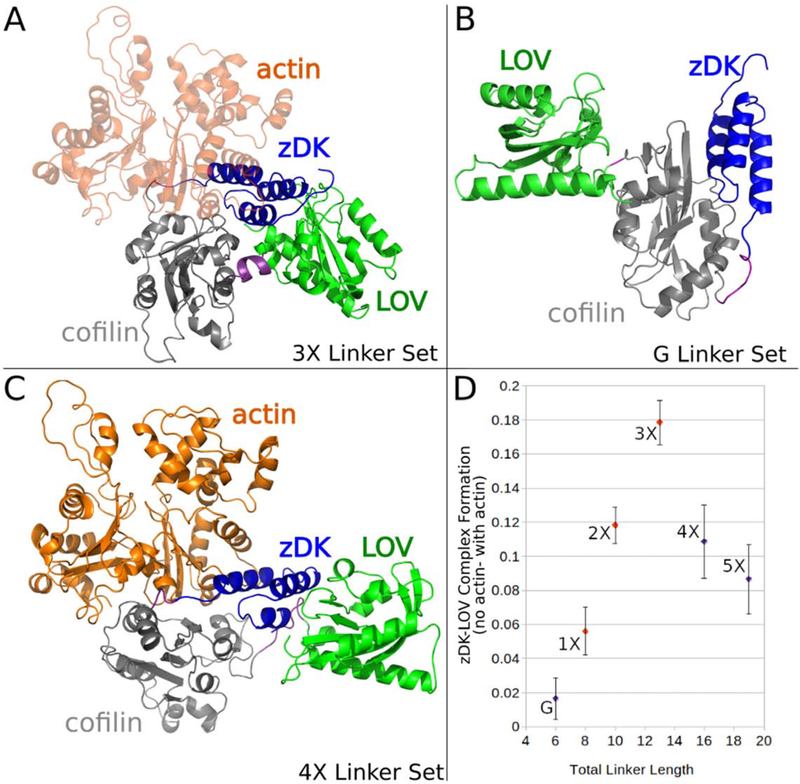
Figure 2.
Figure 2. Modeling linkers for Z-lock cofilin. Various linkers were modeled in the presence and absence of actin to establish which linkers should most effectively regulate binding to cofilin. (a) The optimal-length linker set “3X” positioned the zDK(blue)/LOV(green) interface over cofilin (grey) such that zDK sterically clashed with actin (orange.) (b) The shorter linker set “G” did not permit the formation of a zDK/LOV2 interface even in the monomer state (the pictured model is with the constraint term downweighted to 0 to emulate the predicted behavior of this construct in cells). (c) The longer linker sets permitted the simultaneous formation of an actin-cofilin interface and a zDK/LOV2 interface. (d) The fraction of modeling trajectories that resulted in a well-formed interface between zDK and LOV2 was calculated for simulations with and without actin and the difference of these two fractions was plotted to establish the linker length most likely to inhibit cofilin binding in the dark. The linker length predicted to be most effective (3X) is in good agreement with experimental results that indicated that the 3X showed less binding to actin in the dark than the 2X and 1X linker sets. As expected, the dark-state efficacy of the linkers decreased sharply with decreasing length, a trend that continued with the short linker (blue, “G”), and the longer linkers (4X and 5X) exhibited a more gradual decline in predicted efficacy.