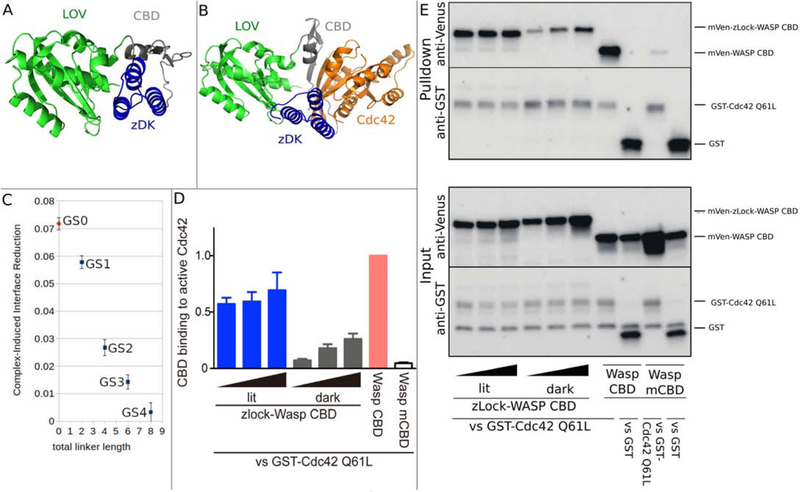
Figure 4.
Computational design and experimental testing of a light-sensitive binder for Cdc42. zDK2(blue) was fused to the N-terminus of the Cdc42 binding motif from Wasp (Wasp CBD, grey) and the LOV2 domain was fused to the C-terminus of Wasp CBD. Simulations were performed in the presence and absence of Cdc42 (orange). (A) In the absence of Cdc42, the n-terminal beta strand of the CBD domain (grey) was allowed to adopt alternative conformations which allowed the the zDK2/LOV2 interaction to form with short linkers. The model shown (GS0) is a direct fusion with no linker residues. (B) With the GS0 linker set, zDK2 is not able to reach the LOV2 domain when the CDB is bound to Cdc42, indicating that Cdc42 binding and zDK2/LOV2 formation will be in competition and that this should be an effective switch. (C) Simulations with longer linkers (GS1-GS4) indicates that caging will be diminished as the linkers are lengthened. (D/E) Experimental analysis of the affinity of dark state and lit state variants of zDK2-GS0-CBD-LOV2 for Cdc42 as determined by pulldown experiments. In panel D the relative amounts of the switch that bound to Cdc42 are quantified for different expression levels. The top portion of panel E shows western blots used to detect the amount of protein in the pulldown and the bottom portion of panel E shows western blots used to detect the total amount of protein in the cell lysate. Lower binding is evident with the dark state variant (top left of panel E).