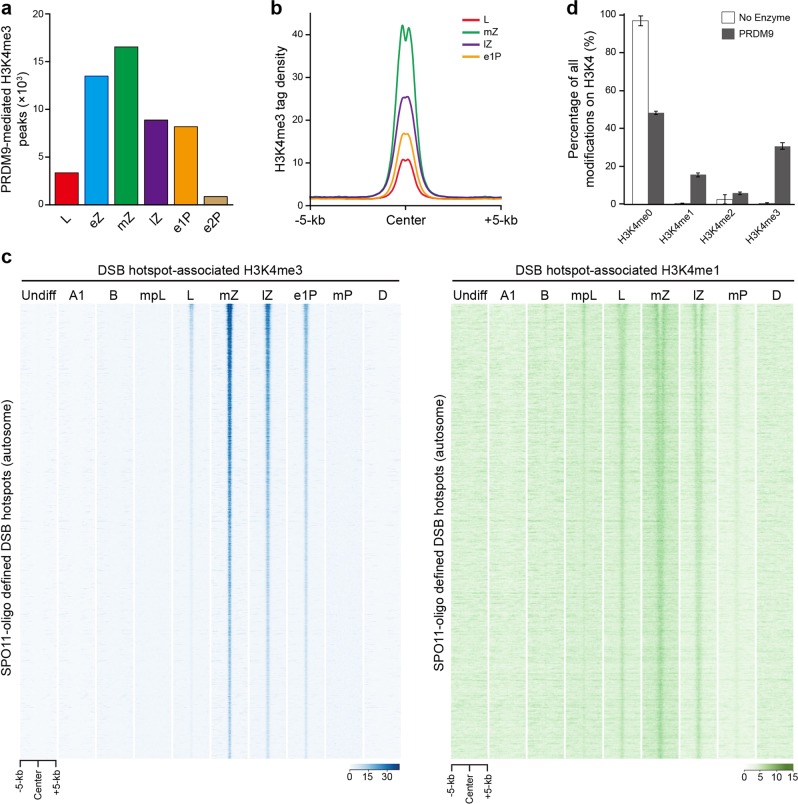
Fig. 2. The dynamics of PRDM9-mediated H3K4me3 marks during progression of recombination.
a Numbers of PRDM9-mediated H3K4me3 peaks in leptotene (L), early-zygotene (eZ), mid-zygotene (mZ), late-zygotene (lZ), early 1-pachytene (e1P), early 2-pachytene (e2P), and mid-pachytene (mP) spermatocytes. b Profile of the averaged H3K4me3 tag density on early-forming hotspot-associated H3K4me3 peaks in leptotene, mid-zygotene, late-zygotene, and early 1-pachytene. H3K4me3 tag density was calculated using H3K4me3 ChIP-seq read coverage with 50-bp resolution. c Heatmaps of H3K4me3 (left) and H3K4me1 (right) normalized tag density on hotspot-associated H3K4me3 peaks in premeiotic and meiotic cells. Each row represents a hotspot-associated H3K4me3 peak of ±5 kb around the peak center and ranked based on SPO11-oligo density from the highest to the lowest. d Histone Methyltransferase Assay using recombinant Prdm9 and oligonucleosomes. The percentage of each methylation state on H3K4 site was quantified by histone mass spectrometry. The blank box represents no enzyme control. Undiff undifferentiated spermatogonia, A1 type A1 spermatogonia, B type B spermatogonia, mpL mid-preleptotene spermatocytes, L leptotene spermatocytes, mZ mid-zygotene spermatocytes, lZ late-zygotene spermatocytes, e1P early 1-pachytene spermatocytes, mP mid-pachytene spermatocytes, D diplotene spermatocytes.