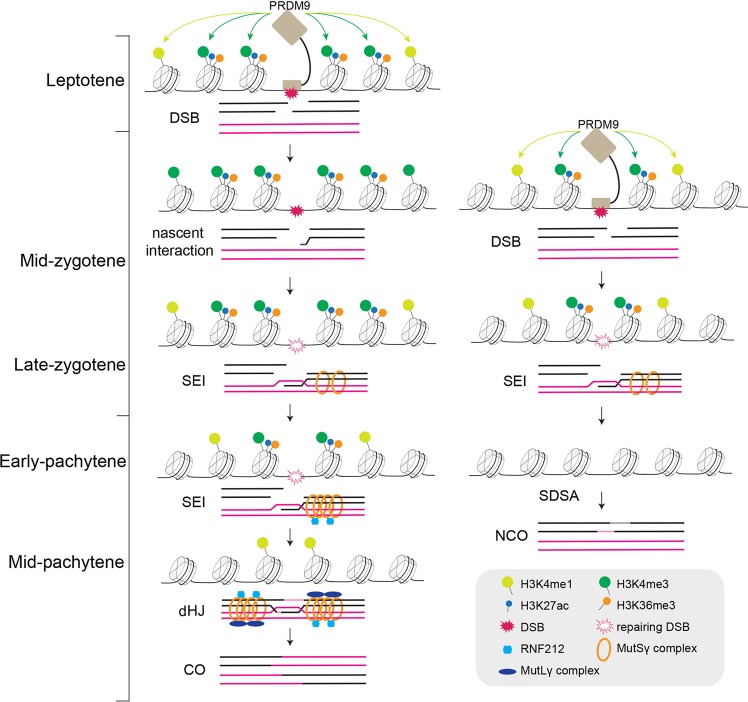
Fig. 6. Proposed model for the DSB fate decision.
At an earlier stage, e.g., leptotene, PRDM9 binds to its target sites containing a longer and stronger binding motif, where it catalyzes the deposition of stronger and extended H3K4me3, and probably directs the deposition of H3K36me3 and H3K27ac. These stronger and more extended H3K4me3 marks may create a stable and permissive chromatin “niche” to predesignate DSB sites to follow a CO fate that facilitates the recruitment of pro-crossover proteins. In contrast, at a later stage, PRDM9 drives deposition of weaker and narrower H3K4me3 marks around its binding sites. The DSBs at the sites of these weaker and narrower H3K4me3 modifications are more likely repaired as NCOs.