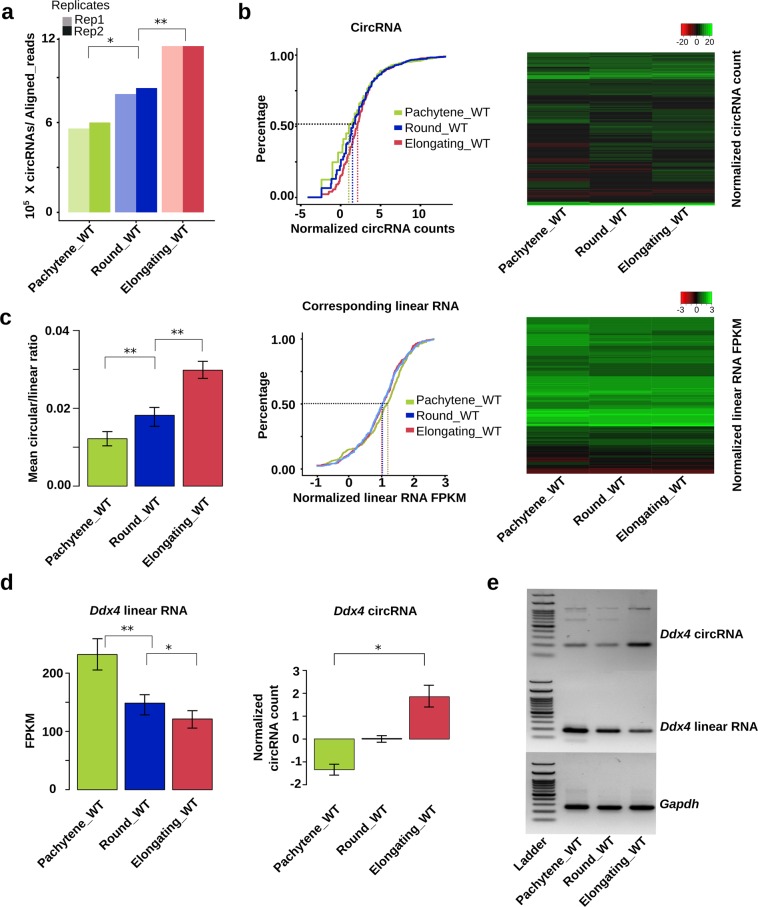
Fig. 1. CircRNA levels increase with progression of spermiogenesis.
a CircRNA expression in wild type (WT) pachytene spermatocytes, round, and elongating spermatids. CircRNA reads were identified from RNA-seq data, followed by normalization against aligned total RNA reads. b CircRNA levels increase while levels of their linear isoforms decrease when WT pachytene spermatocytes develop to round and elongating spermatids. Accumulation curves and heat maps showing normalized circRNA counts and normalized linear RNA levels (FPKM) are used to compare levels of circRNAs and their linear isoforms in WT pachytene spermatocytes, round, and elongating spermatids. c The circular/corresponding linear RNA ratio increases from WT pachytene spermatocytes, to round and elongating spermatids. Normalized counts of circRNAs and their linear isoforms are used to calculate the ratios and to generate the bar graphs. d Ddx4 linear and circRNAs display opposite expression patterns in WT pachytene spermatocytes, round, and elongating spermatids. Normalized circRNA counts and normalized linear RNA levels (FPKM) are compared in WT pachytene spermatocytes, round, and elongating spermatids. e A representative result of semi-quantitative qPCR analyses for levels of Ddx4 circular and linear RNAs in WT pachytene spermatocytes, round, and elongating spermatids. Gapdh was used as a loading control. The Student’s t test was used for statistical analyses. *P < 0.1; **P < 0.05; ***P < 0.001. The data were presented as two biological replicates or means ± SD, biological replicates, n = 2. Sums of two technical repeats in each of the biological replicates were used for analyses.