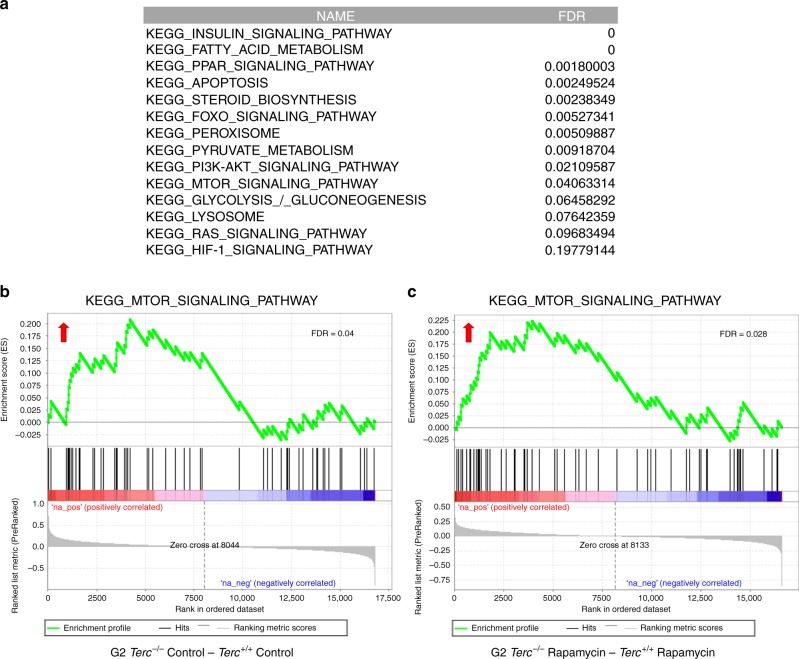
Fig. 5. Telomerase-deficient mice show an upregulated mTOR pathway.
Gene expression data obtained by RNAseq of three independent liver samples from Terc+/+ and G2 Terc−/− male mice subjected to control or rapamycin diet and sacrificed after 2 months of treatment were analyzed by Gene Set Enrichment Analysis (GSEA) to determine significantly enriched gene sets. a Table showing significantly enriched gene sets (FDR < 0.25) between control-fed G2 Terc−/− and Terc+/+. Note the upregulation of several metabolic pathways in telomerase deficient as compared to wild-type mice (for a complete list of deregulated pathways see Supplementary Table 1). The database used was the Kyoto Encyclopedia of Genes and Genomes (KEGG). b, c Gene set enrichment analysis (GSEA) plots for the mTOR pathway in G2 Terc−/− versus Terc+/+ mice subjected to control (b) or rapamycin (c) diet. The red to blue horizontal bar represents the ranked list. Genes located at the central area of the bar show small differences in gene expression between the pairwise compared. At the red edge of the bar are located genes showing higher expression levels in G2 Terc−/− control fed (b) or G2 Terc−/− rapamycin-fed (c) mice; at the blue edge of the bar are located genes showing higher expression levels in Terc+/+ control fed (b) or Terc+/+ rapamycin-fed (c) mice. Red arrows indicated upregulation of the mTOR pathway in the pairwise comparisons. False discovery rate (FDR) is indicated in each case.