Figure 1. Structure of the Plasmodium nth gene and gene product.
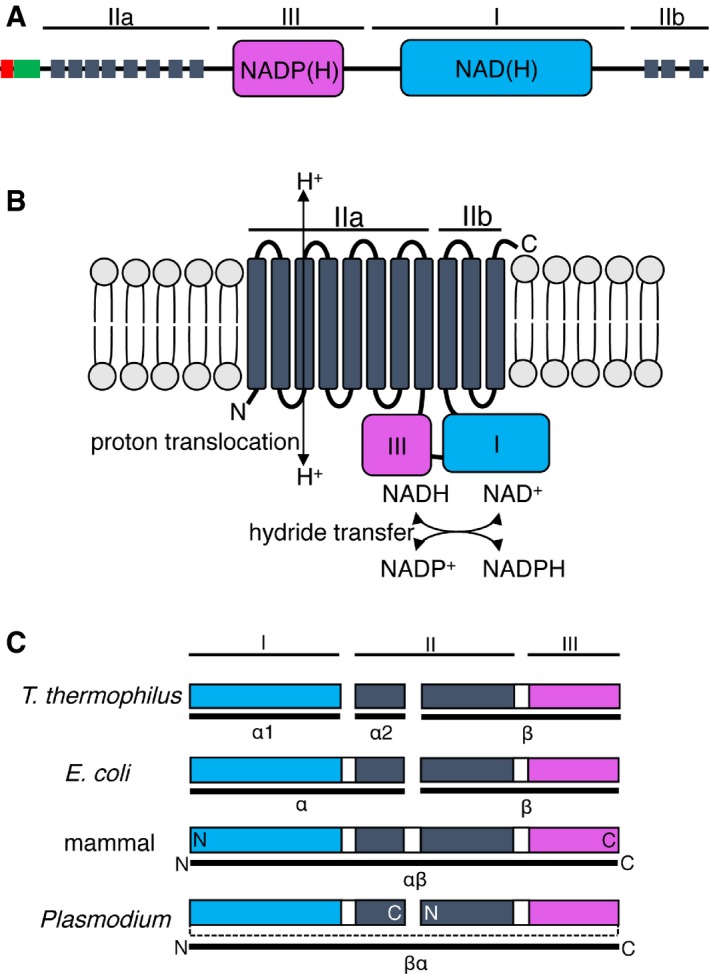
- Schematic domain composition of PBANKA_1317200, showing predicted ER signal peptide (red), apicoplast transit peptide (green), transmembrane helices (dark grey), NADP(H) binding (pink), and NAD(H) binding (blue) modules. Functional NTH domains I, II, and III are indicated.
- Predicted functions of the Plasmodium berghei NTH protomer in the lipid bilayer showing functional domains I–III and corresponding functional activities.
- Organisation of functional NTH domains I–III (coloured bars) in Thermus thermophilus, Escherichia coli, mammals, and Plasmodium. Linker connecting functional domains is indicated by dotted line. Prokaryotic nth genes are segmented into three (T. thermophilus) or two (E. coli) segments, encoding NTH subunits α1, α2, and β (T. thermophilus) or α and β (E. coli). Eukaryotic nth genes are unsegmented and encode single polypeptide NTH of different orientations, either corresponding to an αβ linkage (mammal) or to a βα linkage (Plasmodium). The amino (N) and carboxy (C) termini of the eukaryotic NTH polypeptides are indicated.
