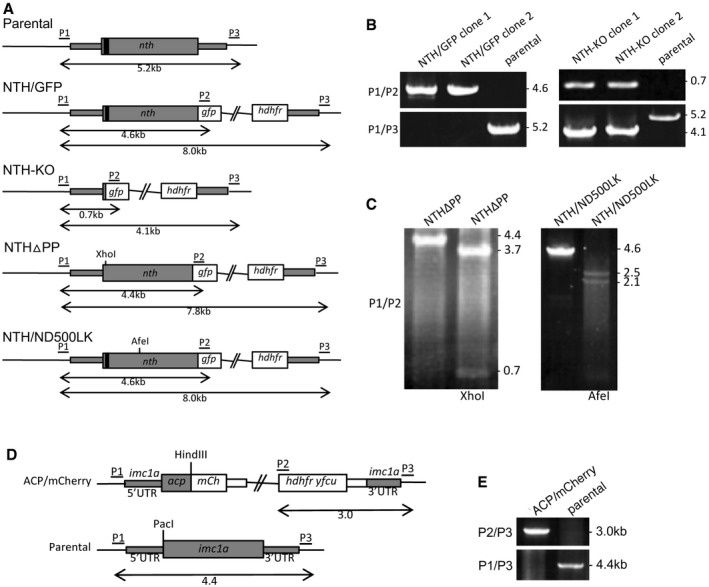
Schematic diagram of the unmodified (parental) and modified nth alleles in parasite lines NTH/GFP, NTH‐KO, NTHΔPP, and NTH/ND500LK. The nth gene is indicated with coding sequence (wide grey bars, introns not shown) and 5′ and 3′ untranslated regions (UTRs; narrow grey bars). Also indicated are the relative positions of the putative apicoplast targeting signal (black box), GFP module (gfp); the human DHFH selectable marker gene cassette (hdhfr); the XhoI and AfeI diagnostic restriction sites; and primers used for diagnostic PCR amplification (P1–P4). Sizes of PCR products are also indicated.
Diagnostic PCR of parasite lines NTH/GFP and NTH‐KO. Integration of the modified GFP‐tagged alleles into the target locus across the 5′ integration site was carried out with primers P1 (ATATTTCCCCCTAATTTTCCCTT) and P2 (GTGCCCATTAACATCACC), amplifying expected products of ˜ 4.6 kb in NTH/GFP and 0.7 kb in NTH‐KO. Absence of the unmodified nth allele was carried out with primers P1 (ATATTTCCCCCTAATTTTCCCTT) and P3 (ATTTCAATACTCGAATTTATGTTATCG), amplifying expected products of ˜ 5.2 kb from the parental parasite and 4.1 kb in NTH‐KO parasites.
Diagnostic PCR and restriction enzyme digests of parasite lines NTHΔPP and NTH/ND500LK. Primers P1 and P2 amplify a 4.4 kb product in NTHΔPP that is digested by XhoI into the expected two fragments of ˜ 3.7 kb and 0.7 kb, confirming presence of the mutation. Primers P1 and P2 amplify a 4.6 kb product in NTH/ND500LK that is digested by AfeI into the expected two fragments of ˜ 2.5 kb and 2.1 kb, confirming presence of the mutation.
Schematic diagram of the modified acp allele (PBANKA_0305600) in parasite line ACP/mCherry. The acp gene is indicated with coding sequence (wide grey bars, introns not shown) and imc1a 5′ and 3′ untranslated regions (UTRs; narrow grey bars). Also indicated are the relative positions of the mCherry module (mCh); the human DHFR::yFCU selectable marker gene cassette (hdhfr yfcu); and primers used for diagnostic PCR amplification (P1–P3). Sizes of PCR products are also indicated.
Diagnostic PCR for integration into the target imc1a locus with primers P2 and P3, giving rise to a 3.0 kb product (top panel). Diagnostic PCR with primer pair P1 and P3 amplified an ˜ 4.4 kb fragment from the parental (wild‐type) parasites, and no product in the ACP/mCherry parasites, confirming absence of the unmodified imc1a allele in the clonal transgenic parasite line (bottom panel).