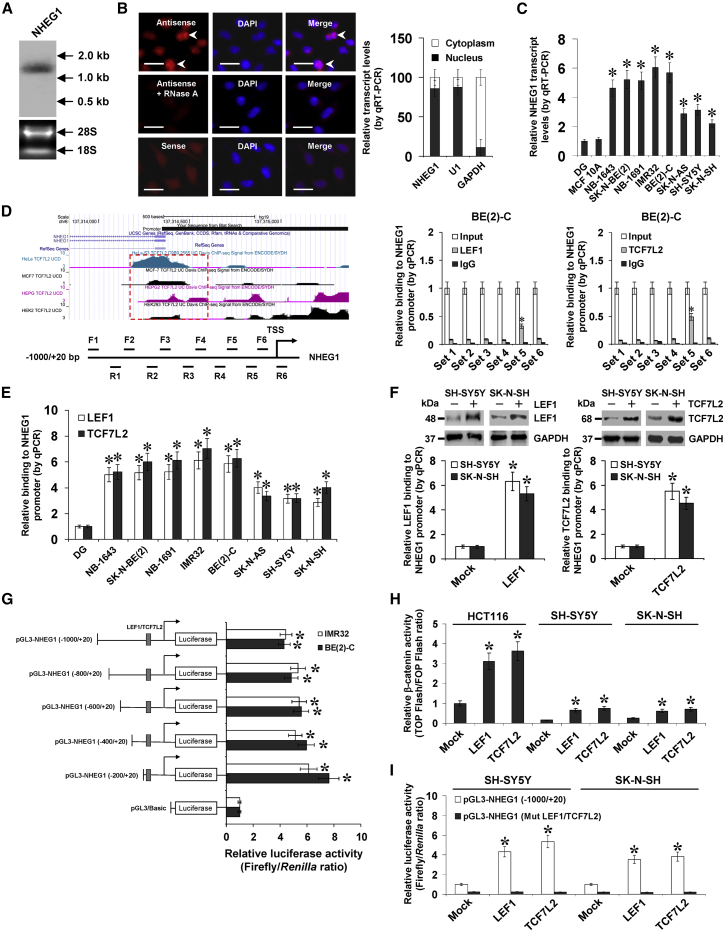
Figure 2.
NHEG1 Is a LEF1/TCF7L2-Regulated Nuclear lncRNA
(A) Northern blot assay with a 268-bp specific probe indicating the existence of 1.3 kb NHEG1 transcript in BE(2)-C cells. (B) RNA fluorescence in situ hybridization images (left panel) showing the nuclear localization of NHEG1 in BE(2)-C cells using a 268-bp antisense probe (red), with nuclei staining with DAPI (blue). The sense probe and antisense probe with RNase A (20 μg) treatment were used as negative controls. Scale bars, 10 μm. Quantitative real-time RT–PCR (right panel) showing the distribution of NHEG1, U1, and GAPDH in the nuclear and cytoplasmic fractions (n = 4). (C) Quantitative real-time RT-PCR assay indicating the NHEG1 transcript levels (normalized to GAPDH) in normal DG (n = 21), MCF 10A cells, and NB cell lines. (D) UCSC Genome Browser view (top left panel) and ChIP and qPCR assays (middle and right panels) with tiled primer sets (bottom left panel), indicating the endogenous binding of TCF7L2 and LEF1 to the NHEG1 promoter (normalized to input DNA) in NB cells (n = 6). (E) ChIP and qPCR assays with primer set 5 showing the enrichment of LEF1 and TCF7L2 on the NHEG1 promoter (normalized to input DNA) in normal DG (n = 10) and NB cell lines (n = 5). (F) ChIP and qPCR assays with primer set 5 (lower panel) and western blot (upper panel) revealing the enrichment of LEF1 and TCF7L2 on the NHEG1 promoter (normalized to input DNA) in NB cells transfected with empty vector (Mock), LEF1, or TCF7L2 (n = 5). (G) Dual-luciferase assay indicating the activity of NHEG1 promoter reporters (normalized to pGL3-Basic) in NB cells (n = 6). (H) Dual-luciferase assay showing the relative activity of β-catenin in colon cancer HCT116 cells and NB cell lines transfected with mock, LEF1, or TCF7L2 (n = 5). (I) Dual-luciferase assay indicating the relative activity of the NHEG1 promoter reporter pGL3-NHEG1 (−1,000/+20) with the wild-type (WT) or mutant (Mut) LEF1/TCF7L2 binding site in NB cells transfected with mock, LEF1, or TCF7L2 (n = 5), with normalization to activity of WT reporter in mock cells. ANOVA and Student’s t test compared the difference in (C)–(I). *p < 0.01 versus DG, IgG, mock, or pGL3-Basic. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (B)–(I).