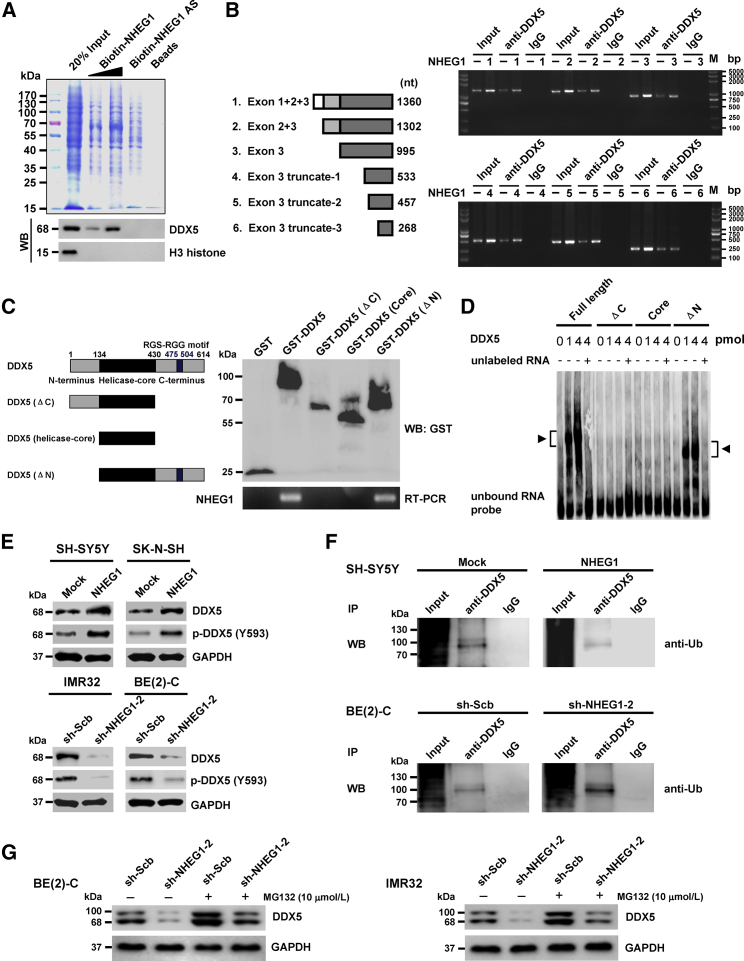
Figure 4.
NHEG1 Interacts with and Stabilizes DDX5 Protein in NB Cells
(A) Biotin-labeled RNA pull-down, Coomassie blue staining (top panel) and validating western blot (bottom) assays revealing the interaction between NHEG1 and DDX5 protein in BE(2)-C cells. NHEG1 antisense (AS)- and bead-bound protein served as negative controls. (B) RIP assay using DDX5 antibody indicating the interaction between NHEG1 and DDX5 protein in SH-SY5Y cells transfected with a series of NHEG1 truncations. The immunoglobulin G (IgG)-bound RNA was taken as negative control. (C) In vitro binding assay depicting the recovered NHEG1 levels by RIP (right lower panel) after incubation with full-length (1–614 amino acids), ΔC (1–430 amino acids), helicase-core (135–430 amino acids), or ΔN (135–614 amino acids) of GST-tagged recombinant DDX5 protein (left panel) validated by western blot (right upper panel). (D) RNA EMSA assay determining the interaction between recombinant DDX5 protein and biotin-labeled RNA probe for NHEG1 (arrowheads), with or without competition, using an excess of the unlabeled homologous RNA probe. (E) Western blot assay showing the total and phosphorylated (at tyrosine 593) DDX5 in NB cells stably transfected with empty vector (Mock), NHEG1, scramble shRNA (sh-Scb), or sh-NHEG1-2. (F) coIP and western blot assays indicating the endogenous interaction between DDX5 and ubiquitin (Ub) in SH-SY5Y and BE(2)-C cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2. (G) Western blot assay showing the expression of DDX5 in NB cells stably transfected with sh-Scb or sh-NHEG1-2 and those treated with proteasome inhibitor MG132 for 48 hr. Data are representative of three independent experiments in (A)–(G).