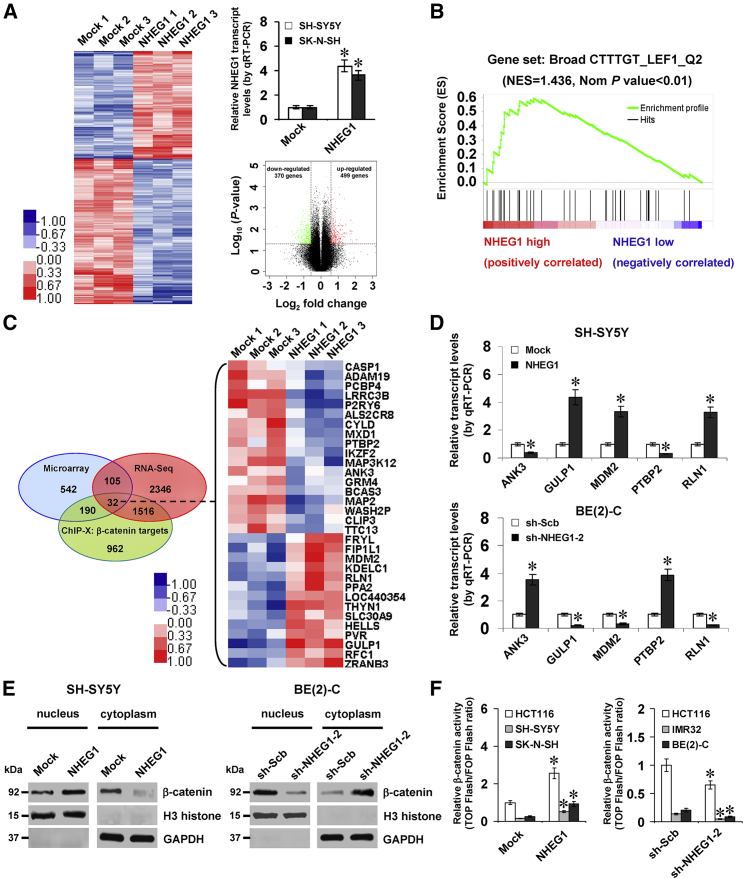
Figure 5.
NHEG1 Regulates the Expression of β-Catenin Target Genes in NB Cells
(A) Microarray heatmap (left, fold change > 1.5, p < 0.05), quantitative real-time RT-PCR (top right panel, n = 3), and volcano plots (bottom right panel) revealing the alteration of gene expression in SH-SY5Y cells stably transfected with empty vector (Mock) or NHEG1. (B) Gene set enrichment analysis of altered genes in SH-SY5Y cells stably transfected with NHEG1, compared with mock-transfected control. NES, normalized enrichment score; Nom, normalized. (C) Venn diagram (left panel) and heatmap (right panel, fold change > 1.5, p < 0.05) showing the NHEG1-altered expression of β-catenin target genes that were intersected with NHEG1-correlated transcripts in the RNA-seq dataset (GEO: GSE62564) and ChIP-X database. (D) Quantitative real-time RT-PCR assay indicating the transcript levels of β-catenin target genes ANK3, GULP1, MDM2, PTBP2, and RLN1 (normalized to GAPDH) in NB cells stably transfected with mock, NHEG1, scramble shRNA (sh-Scb), or sh-NHEG1-2 (n = 5). (E) Western blot assay showing the nuclear and cytoplasmic expression of β-catenin and H3 histone in NB cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2. (F) Dual-luciferase assay indicating the relative activity of β-catenin in colon cancer HCT116 and NB cells transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2 (n = 5). Student’s t test analyzed the difference in (A), (D), and (F). Fisher’s exact test for overlapping analysis in (C). *p < 0.01 versus mock or sh-Scb. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (D)–(F).