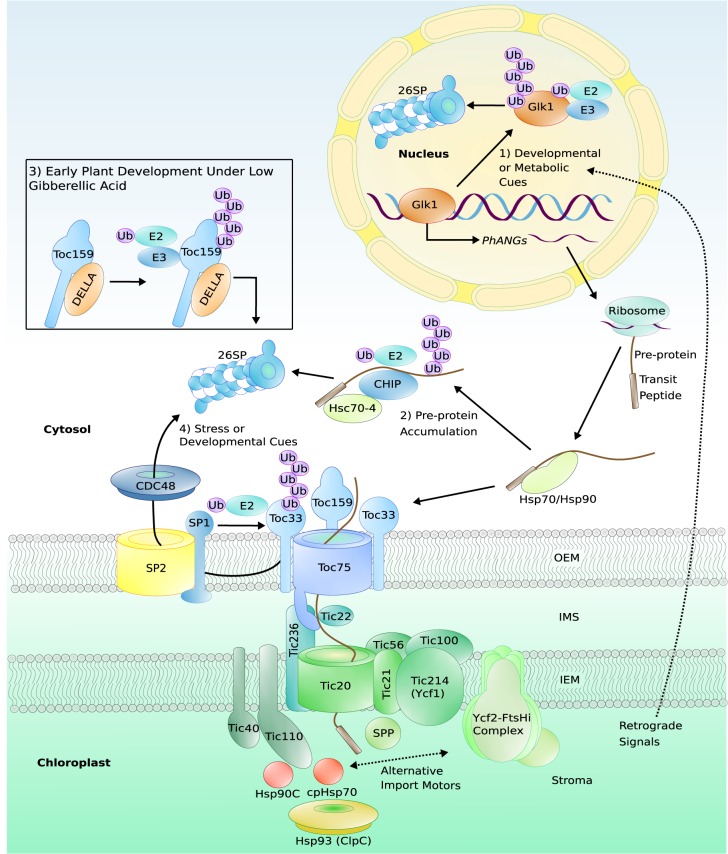
Figure 1. An overview of protein import into the chloroplast and its regulation by the UPS.
Nucleus-encoded chloroplast proteins are synthesized in the cytosol as pre-proteins and post-translationally imported into the chloroplast. The pre-protein carries an N-terminal transit peptide which holds guidance information and initially allows interaction with cytosolic chaperones (e.g. Hsp70, Hsp90). The pre-protein is then recognized by receptor GTPases in the OEM, Toc33 and Toc159 (which exist in different isoforms with varying client specificity, and in Arabidopsis, these are termed Toc34 and Toc132/-120/-90, respectively). The receptors heterodimerize to allow the pre-protein to pass through the Toc75 pore into the IMS. Passage through the IEM is mediated by Tic20, which is reported to be part of the 1 MDa TIC complex containing Tic21, Tic56, Tic100, and Tic214 (Ycf1). Completion of translocation into the stroma is powered by an ATP-dependent import motor, which may be composed of stromal molecular chaperones (e.g. cpHsp70, Hsp90C) or a 2 MDa Ycf2–FtsHi complex. The Tic40 and Tic110 proteins are also involved in the import process, and may operate downstream in conjunction with stromal chaperones. Hsp93 (ClpC) has been proposed to perform a quality-control function at the point of import, or to act in the import motor. Upon arrival in the stroma, the transit peptide is cleaved from the pre-protein by the SPP. The UPS regulates protein import in a variety of ways: (1) The transcription factor Glk1, which regulates the expression of pre-protein-encoding PhaNGs, may be degraded by the UPS in response to unknown retrograde signals (from the chloroplast to the nucleus) that report on developmental or metabolic cues. (2) Accumulation of pre-proteins in the cytosol may trigger their UPS degradation to prevent the formation of cytotoxic aggregates, and this is mediated by the chaperone Hsc70-4 and the E3 ligase CHIP. (3) Before germination, DELLA factors repress chloroplast biogenesis under low gibberellic acid conditions by binding to Toc159 and triggering its UPS degradation. (4) During stress or particular phases of development, the CHLORAD system directly targets the TOC apparatus for proteolysis, with ubiquitination being mediated by the E3 ligase SP1. The targeted TOC proteins are retrotranslocated from the membrane via the channel protein SP2, using motive force provided by the cytosolic AAA+ ATPase CDC48. Note that the Toc33 and Toc159 receptor isoforms are depicted in the model here due to the known role of CHLORAD in suppressing the import of photosynthetic pre-proteins in response to abiotic stress [112], and because such photosynthetic pre-proteins are the primary clients of these isoforms [2]; however, all of the TOC receptor isoforms are likely substrates of SP1 at some stage, as revealed for example by the analysis of sp1 plants during de-etiolation [109]. Dashed lines indicate uncertainty. Abbreviations: E2, E2 conjugase; E3, E3 ligase; 26SP, 26S proteasome; Ub, ubiquitin; SP, suppressor of ppi1; CDC48, cell division cycle 48; TOC, translocon at the outer envelope membrane of chloroplasts; TIC, translocon at the inner envelope membrane of chloroplasts; OEM, outer envelope membrane; IMS, intermembrane space; IEM, inner envelope membrane; SPP, stromal processing peptidase; GLK1, Golden2-like1; CHIP, C-terminus of Hsc70-interacting protein; PhANGs, photosynthesis-associated nuclear genes.