Endoplasmic reticulum-localized phosphoinositide phosphatase hydrolyzes phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate to regulate cell elongation in rice.
Abstract
Phosphoinositides (PIs) as regulatory membrane lipids play essential roles in multiple cellular processes. Although the exact molecular targets of PI-dependent modulation remain largely elusive, the effects of disturbed PI metabolism could be employed to identify regulatory modules associated with particular downstream targets of PIs. Here, we identified the role of GRAIN NUMBER AND PLANT HEIGHT1 (GH1), which encodes a suppressor of actin (SAC) domain-containing phosphatase with unknown function in rice (Oryza sativa). Endoplasmic reticulum-localized GH1 specifically dephosphorylated and hydrolyzed phosphatidylinositol 4-phosphate (PI4P) and phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. Inactivation of GH1 resulted in massive accumulation of both PI4P and PI(4,5)P2, while excessive GH1 caused their depletion. Notably, superabundant PI4P and PI(4,5)P2 could both disrupt actin cytoskeleton organization and suppress cell elongation. Interestingly, both PI4P and PI(4,5)P2 inhibited actin-related protein2 and -3 (Arp2/3) complex-nucleated actin-branching networks in vitro, whereas PI(4,5)P2 showed more dramatic effects in a dose-dependent manner. Overall, the overaccumulation of PI(4,5)P2 resulting from dysfunction of SAC phosphatase possibly perturbs Arp2/3 complex-mediated actin polymerization, thereby disordering cell development. These findings imply that the Arp2/3 complex might be the potential molecular target of PI(4,5)P2-dependent modulation in eukaryotes, thereby providing insights into the relationship between PI homeostasis and plant growth and development.
Phosphoinositides (PIs) as a minor component of cell membranes and phosphorylated derivatives of phosphatidylinositol liberate the second messengers, inositol 1,4,5-triphosphate and diacylglycerol, in response to activation of cell surface receptors (Boss and Im, 2012). Nevertheless, emerging evidence suggests that PIs also play crucial roles as signaling molecules, functioning as regulatory lipids in various fundamental cellular processes (Heilmann, 2016). A new detection approach further confirmed that phosphatidylinositol 4-phosphate (PI4P) and phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] are the most abundant PIs in cells (Balla, 2013; Kielkowska et al., 2014). PI4P is phosphorylated by phosphatidylinositol phosphate kinases, resulting in the production of PI(4,5)P2, while conversely, PI(4,5)P2 is dephosphorylated by PI phosphatases to generate PI4P, supporting the hypothesis that turnover and dynamic formation of PIs is necessary for plant development (Gerth et al., 2017).
The plant actin cytoskeleton plays essential roles in cell morphogenesis, and the dynamics of the actin cytoskeleton drive membrane deformation and trafficking, which is required for polar cell growth and movement of vesicles and organelles (Pleskot et al., 2014; Wang et al., 2017). This dynamic remodeling is controlled by diverse actin-binding proteins, which underpin various signaling pathways. Membrane PIs also affect the dynamic assembly and disassembly of actin filaments (Li et al., 2015; Gerth et al., 2017); notably, PI(4,5)P2 regulates the activity and distribution of actin-binding proteins, which in turn are tightly linked to actin polymerization activity (Saarikangas et al., 2010; Zhang et al., 2012; Bothe et al., 2014; Pleskot et al., 2014).
Moreover, the highly conserved actin-related protein2 (Arp2) and Arp3 complex was found to nucleate branched actin filament networks, although it is dependent on the stimulation of nucleation-promoting factors (NPFs) such as Wiskott-Aldrich syndrome protein (WASP) and WASP family verprolin homologous-protein (WAVE)/suppressor of cAMP receptor (SCAR; Pollard, 2007; Rotty et al., 2013). NPFs tend to share a typical C-terminal tripartite functional VCA (verprolin homology, central hydrophobic, and acidic C-terminal) domain, which requires PI(4,5)P2 to overcome autoinhibition and bind to the Arp2/3 complex for conformation activation (Pollard, 2007; Padrick and Rosen, 2010). In plants, activation of the Arp2/3 complex is relatively simple and appears dependent on the WAVE/SCAR family and the heteromeric WAVE/SCAR regulatory complex as a sole trigger (Frank et al., 2004; Yanagisawa et al., 2013). Nevertheless, the precise cellular control of WAVE/SCAR-Arp2/3 module-nucleated actin polymerization and the relationship with membrane PIs remains unknown.
In general, the reversible phosphorylation of PIs dominated by a set of kinases and phosphatases controls the turnover and dynamics of membrane PIs (Hsu and Mao, 2013; Takasuga and Sasaki, 2013). PI phosphatases such as inositol polyphosphate phosphatases and suppressor of actin (SAC) domain-containing phosphatases were also found to hydrolyze phosphate from PIs, thereby modifying PI levels (Zhong and Ye, 2003; Ercetin and Gillaspy, 2004; Zhong et al., 2005; Thole et al., 2008; Nováková et al., 2014; Gerth et al., 2017). However, the cellular functions of the SAC phosphatases involved in PI metabolism during plant growth and development remain largely unknown in crops. This study carried out functional characterization of an unknown SAC phosphatase in rice (Oryza sativa) that is required for homeostasis of PI(4,5)P2 and PI4P and thus affects plant cell morphogenesis. These results indicate that the overaccumulation of PI(4,5)P2 possibly disrupts the Arp2/3 complex activity and actin cytoskeleton organization, thereby suppressing cell elongation in rice.
RESULTS
GH1 Contributes to Rice Plant and Panicle Development
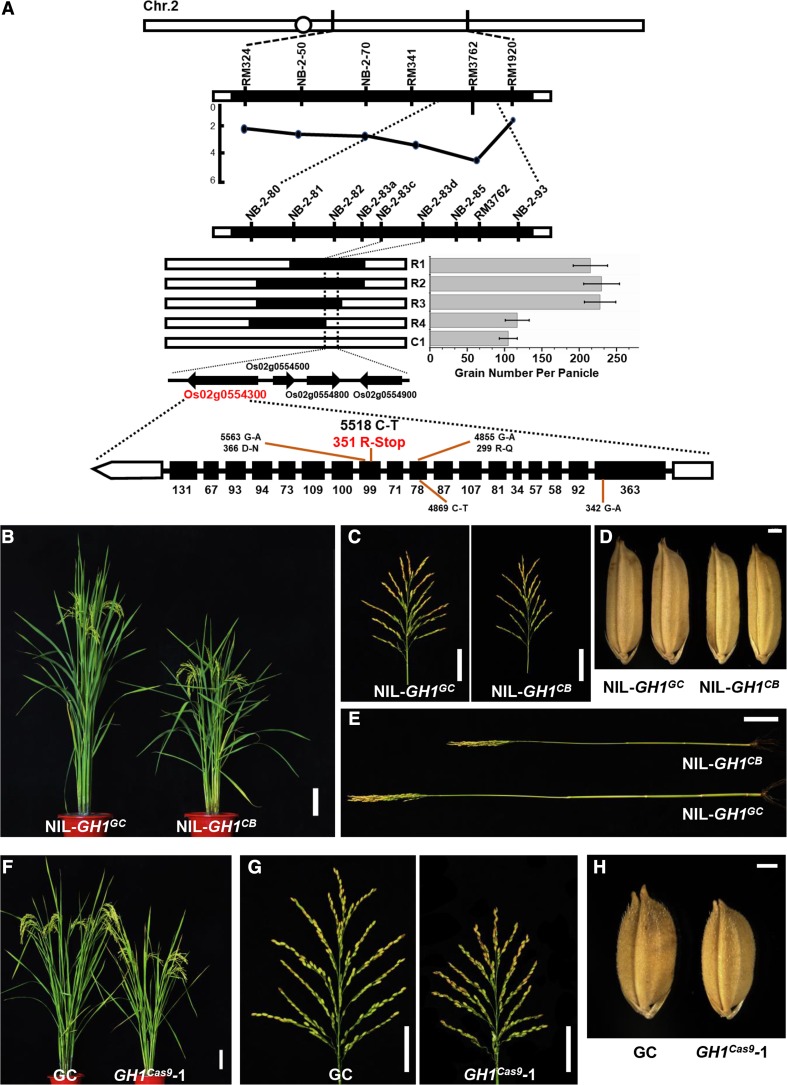
To determine the molecular basis of panicle morphogenesis in rice, parental varieties Guichao-2 (GC) and CB, which exhibit obvious differences in plant and panicle architecture, were selected to identify new quantitative trait loci (Supplemental Fig. S1, A–H). GC and CB were crossed to produce an F2 mapping population, and then a map-based cloning method was used to isolate a new gene locus for grain number per panicle located in the region of chromosome 2 defined using molecular markers NB-2-80 and NB-2-93 (Fig. 1A). These findings coincide with the results of bulked segregant analysis-assisted mapping (Supplemental Fig. S2; Takagi et al., 2013). Therefore, these results suggest that a single locus, referred to here as GRAIN NUMBER AND PLANT HEIGHT1 (GH1), is pleiotropically responsible for plant and panicle morphogenesis. The GH1 gene was then fine-mapped to a region between marker loci NB-2-83c and NB-2-83d using a larger segregating F2 population (Fig. 1A). This region contained four predicted genes, but sequence comparisons using the region from the mapping parents showed that only Os02g0554300 was polymorphic. It was therefore selected as the candidate gene of GH1, in which a C-to-T nucleotide mutation from the CB genotype resulted in a premature stop codon of this protein (Fig. 1A).
Figure 1.
GH1 contributes to rice plant and panicle development. A, Map-based cloning of GH1. Grain number per panicle of the three recombinant lines (R1–R3) was higher than that of recombinant line R4 and the control (C1, homozygous for CB in the target region; n = 15 plants). Values represent means ± sd. Black and white bars represent homozygous chromosomal segments for GC and CB, respectively. Four open reading frames were found in the candidate region, Os02g0554300, Os02g0554500, Os02g0554800, and Os02g0554900. Sequencing of the full length of the four genes revealed a C-T mutation in the 11th exon of Os02g0554300, resulting in premature stop of this protein. B, Plant architecture of NIL-GH1GC and NIL-GH1CB at the reproductive phase. Bar = 10 cm. C, Mature panicles of NIL-GH1GC and NIL-GH1CB. Bars = 5 cm. D, Mature grains from NIL-GH1GC and NIL-GH1CB. Bar = 1 mm. E, The culms of NIL-GH1GC and NIL-GH1CB. Bar = 5 cm. F, Plant architecture of GC and GH1cas9-1 at the reproductive phase. Bar = 10 cm. G, Mature panicles of GC and GH1cas9-1. Bars = 5 cm. H, Mature grains of GC and GH1cas9-1. Bar = 1 mm.
To estimate the effects of GH1, a nearly isogenic line (NIL) containing a 50-kb chromosomal region of GC at the GH1 locus was developed under the CB genetic background. NIL-GH1CB displayed a distinct short stem length and reduced panicle and grain size compared with NIL-GH1GC (Fig. 1, B–E). Accordingly, the average plant height, grain number per panicle, and grain length and width of NIL-GH1CB decreased significantly (Supplemental Fig. S1, I–L). The identity of GH1 was then validated by editing the candidate gene using the CRISPR/Cas9 method (Supplemental Fig. S3; Ma et al., 2015), revealing a similar phenotype between GH1 knockout line GH1cas9 and NIL-GH1CB, which had smaller panicles and grains and was shorter in height (Fig. 1, F–H; Supplemental Fig. S4, A–G and J–M). Overexpression of GH1 also resulted in weaker growth of plant and panicle architecture compared with the wild type (Supplemental Fig. S5, A–H). Overall, these results suggest that GH1 plays a crucial role in plant and panicle development during rice morphogenesis.
GH1 Phosphatase Specifically Dephosphorylates PI4P and PI(4,5)P2
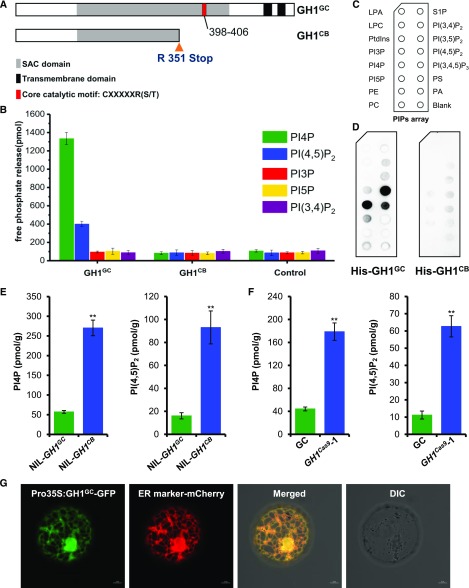
GH1 was therefore thought to encode a previously unknown SAC domain-containing phosphatase in rice that is conserved throughout the plant kingdom (Supplemental Fig. S6). Notably, mutation of Arg at position 351 directly resulted in premature stop of GH1CB at the CB allele and deletion of the core catalytic motif in SAC phosphatase (Fig. 2A). Recently, SAC domain-containing proteins were found to possess PI phosphatase activity in eukaryotic species (Wei et al., 2003; Liu et al., 2008; Thole et al., 2008; Brice et al., 2009; Lee et al., 2011; Hsu and Mao, 2013). Although the SAC domain is homologous among different proteins, they appear to display varied substrate specificity and subcellular localization (Hsu and Mao, 2013). Nevertheless, the specific functions of SAC phosphatase in crops remain largely unknown. Thus, to elucidate the phosphatase activity and substrate specificity of GH1, GH1GC and GH1CB fusion proteins from Sf9 insect cells were purified for in vitro phosphatase activity assay. Malachite Green phosphate assay was then used to determine the enzyme activity of PI phosphatase. Interestingly, GH1GC protein exhibited preference and substrate specificity toward PI4P and PI(4,5)P2 rather than PI3P, PI5P, and PI(3,4)P2 compared with the control, with a higher level of PI4P phosphatase activity in vitro (Fig. 2B). Moreover, GH1CB failed to recognize the diverse PI isoforms, especially PI4P and PI(4,5)P2, suggesting that mutation of GH1CB disabled its phosphatase activity (Fig. 2B). Furthermore, to test whether GH1 binds to phospholipids directly, the fusion proteins were purified and subjected to a protein-lipid overlay assay on which 15 different lipids were spotted (Fig. 2C). The results showed that the fusion protein GH1GC can strongly bind to PI4P and PI(4,5)P2 and show weak binding to PI(3,4,5)P3; however, the mutated GH1CB hardly binds to the phospholipids (Fig. 2D). These results also coincide with the previous suggestion that the conserved catalytic site of the SAC domain consisting of a peptide motif with a CX5R sequence at the C-terminal end plays an essential role in phosphate hydrolysis in yeast (Saccharomyces cerevisiae; Manford et al., 2010). Overall, these findings suggest that the SAC phosphatase GH1 specifically dephosphorylates and hydrolyzes PI4P and PI(4,5)P2.
Figure 2.
GH1 phosphatase specifically dephosphorylates PI4P and PI(4,5)P2. A, C-T mutation of the GH1 allele from CB caused a premature stop codon that resulted in absence of the core catalytic motif and two transmembrane domains in the C-terminal end of the predicted SAC domain-containing phosphatase. B, The SAC domain-containing GH1GC phosphatase showed high substrate specificity toward PI4P and PI(4,5)P2 over other PI isoforms, while the mutated GH1CB showed no phosphatase activity as a control (n = 3 biologically independent reactions). Values represent means ± sd. C, Schematic diagram of a PIP strip containing an array of immobilized phospholipids: lysophosphatidic acid (LPA), lysophosphocholine (LPC), phosphatidylinositol (PtdIns), PI3P, PI4P, PI5P, phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine-1-phosphate (S1P), PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, PI(3,4,5)P3, phosphatidylserine (PS), and phosphatidic acid (PA). D, Purified recombinant His-GH1GC (left) and His-GH1CB (right) were overlaid onto PIP strip membranes. Proteins bound to lipids were detected by immunoblotting with anti-His monoclonal antibody. E and F, Comparisons of endogenous levels of PI4P and PI(4,5)P2 extracted from rice leaf blades of NIL-GH1GC and NIL-GH1CB (E) and GC and GH1cas9-1 plants (F; n = 3 biologically independent assays). Values represent means ± sd. **, P < 0.01 compared with the control using Student’s t test. G, Subcellular localization of GH1 in rice protoplasts. The GH1 protein was shown to target the ER by transient expression of GH1GC-GFP, which merged with the ER marker mCherry. DIC, Differential interference contrast. Bars = 5 μm.
To further investigate the effect of GH1 on PI4P and PI(4,5)P2 metabolism in rice, endogenous levels of PI4P and PI(4,5)P2 were determined using ELISA with different plant leaves. Both PI4P and PI(4,5)P2 levels were markedly higher in NIL-GH1CB than NIL-GH1GC (Fig. 2E), suggesting that loss of function of GH1 results in massive accumulation of these PI isoforms in rice. Consistent with this, the GH1 knockout line GH1cas9 also showed a significant increase in endogenous PI4P and PI(4,5)P2 compared with the wild type (Fig. 2F; Supplemental Fig. S4, H and I). In contrast, GH1OE overexpressing plants had reduced PI4P and PI(4,5)P2 levels (Supplemental Fig. S5, I and J). These findings suggest that GH1 specifically acts as a PI4P and PI(4,5)P2 phosphatase, maintaining degradation and homeostasis of PI4P and PI(4,5)P2, and thereby acting as an essential regulator of membrane PI signaling networks in rice. Membrane lipids can be transferred between bilayers at contact sites between the endoplasmic reticulum (ER) and other membranes to maintain homeostasis (Stefan et al., 2013; Holthuis and Menon, 2014; Chung et al., 2015). In previous reports, countertransport and exchange of PI4P and PS between the ER and plasma membrane (PM) enabled the delivery of PI4P to the ER during degradation and synthetic PS from the ER to the PM, controlling PI4P levels and selectively enriching PS in the PM (Stefan et al., 2011; Chung et al., 2015). In both yeast and mammalian cells, the PI4P phosphatase Sac1 is localized in the ER and Golgi apparatus, where it acts as an integral membrane protein, and PI4P is hydrolyzed by the ER protein Sac1 (Stefan et al., 2011; Mesmin et al., 2013). These findings suggest that GH1 also might function as an ER protein in rice, with two tandem transmembrane domains in its C-terminal end (Supplemental Fig. S7). Subcellular localization of GH1 in rice protoplasts was therefore examined using fluorescence signals from a GFP (GH1-GFP) fusion protein. Signals coincided with those of the ER marker protein (Nelson et al., 2007), suggesting that the SAC phosphatase GH1 degrades PI4P and PI(4,5)P2 within the ER (Fig. 2G).
GH1 Is Required for Actin Cytoskeleton Organization and Organelle Development in Rice
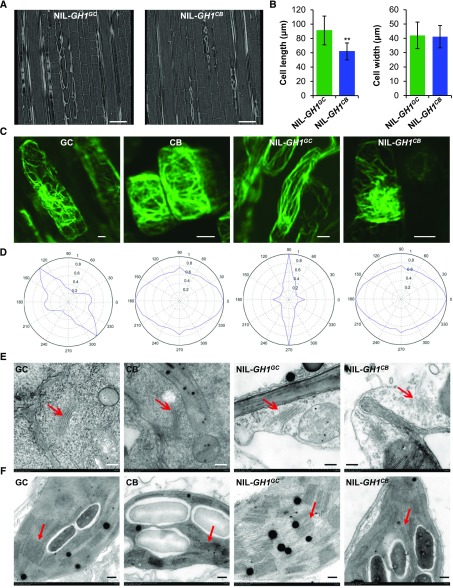
To understand the cellular roles of the SAC phosphatase GH1, vertical sections of the central part of the culm were therefore compared between NIL-GH1GC and NIL-GH1CB using x-ray microscopy to determine the cellular basis for the short stem. Observations revealed that the average cell length of the culm was significantly smaller in NIL-GH1CB than NIL-GH1GC, whereas the cell width was comparable (Fig. 3, A and B). These findings suggest that GH1 contributes to cell morphogenesis in rice, with deletion suppressing cell elongation. It has been suggested that SAC domain-containing proteins influence the organization of the actin cytoskeleton in yeast and plant cells (Foti et al., 2001; Zhong et al., 2005). To explore whether GH1 is also associated with actin cytoskeleton modulation in rice, the organization of actin filaments in the root tip cells was also examined. Root tip cells from parent GC and NIL-GH1GC plants formed a structured actin filament cable network, while in CB and NIL-GH1CB, the actin filaments appeared to have lost their organized structure (Fig. 3C), suggesting that GH1 is responsible for the fine organization of the actin cytoskeleton. Actin filament orientation in all directions (360°) was therefore examined using visual statistical analysis, revealing a specific direction in GC and NIL-GH1GC. Meanwhile, in CB and NIL-GH1CB, the actin cytoskeleton displayed a disordered distribution without definite orientation (Fig. 3D). Additionally, statistical analysis of the filament length and number of filaments showed that the average lengths of actin filaments from CB and NIL-GH1CB were significantly shorter compared with GC and NIL-GH1GC; however, the number of actin filaments in CB and NIL-GH1CB increased (Supplemental Fig. S8, A and B). These results support the idea that CB and NIL-GH1CB could have significantly fragmented actin filaments. Consistent with this, both PI4P and PI(4,5)P2 levels were markedly higher in roots of NIL-GH1CB than NIL-GH1GC (Supplemental Fig. S8C). These findings imply that homeostasis of membrane PI4P and PI(4,5)P2 under GH1 control could be required for actin cytoskeleton organization in rice. Furthermore, the seedlings were observed, and the average root length of NIL-GH1CB was obviously shorter than in NIL-GH1GC (Supplemental Fig. S9, A and C), and the root cells were distinctly smaller (Supplemental Fig. S9, B and D). These results suggested that the elevated PI4P and PI(4,5)P2 induced by malfunction of GH1 could suppress cell development.
Figure 3.
GH1 is required for actin cytoskeleton organization and organelle development in rice. A, Scanning of vertical sections of the rice culm from NIL-GH1GC and NIL-GH1CB using Xradia 510 versa 3D x-ray microscopy. Bars = 100 μm. B, Comparisons of cell length and width within the culm of NIL-GH1GC and NIL-GH1CB (n = 10 views from three plants). Values represent means ± sd. **, P < 0.01 compared with NIL-GH1GC using Student’s t test. C, F-actin observations of rice root tip cells from GC, CB, NIL-GH1GC, and NIL-GH1CB stained with ActinGreen488. More than 10 views from five plants for each allele and at least one to two cells in every view were observed, and representative views are shown for each allele. Bars = 5 μm. D, Visual statistical analysis of actin filament orientation and actin cytoskeleton arrangement in all directions (360°) using Discrete Fourier Transform. In each allele, 10 different and representative subsamples from five plants containing at least 20 cells were analyzed. The most representative radar charts from one cell in each allele are shown. E and F, Observations of Golgi apparatus (E) and chloroplasts (F) from GC, CB, NIL-GH1GC, and NIL-GH1CB using transmission electron microscopy. The red arrows indicate the layered Golgi apparatus and thylakoids, respectively. Bars = 2 μm.
A previous study revealed that lipid phosphatase, in regulating PI homeostasis, plays an essential role in Golgi membrane organization in mammals (Liu et al., 2008). However, whether SAC phosphatases play a similar biological role in plants remains unknown. Golgi morphology in rice leaf cells was therefore examined by transmission electron microscopy. Intact multilayered Golgi apparatuses were observed in the GC and NIL-GH1GC cells; however, in CB and NIL-GH1CB, they were impaired with concomitant collapse and vesicle-like structures (Fig. 3E; Supplemental Fig. S10A). These findings therefore suggest a potential role of GH1 in Golgi development and vesicle trafficking in plants. Moreover, they also suggest that elevated PI4P and PI(4,5)P2 levels could disrupt the structural integrity and function of the Golgi apparatus.
Chloroplast morphology in the leaf cells was subsequently examined, revealing a dramatic reduction in thylakoid layers and the number of grana in CB and NIL-GH1CB compared with GC and NIL-GH1GC (Fig. 3F; Supplemental Fig. S10B). These findings suggest that increased levels of membrane PI4P and PI(4,5)P2 could also disrupt thylakoid formation and grana assembly during chloroplast development in rice. Interestingly, this result coincides with reported evidence whereby inhibition of PI4P synthesis accelerates chloroplast division, supporting the idea that PI4P negatively regulates chloroplast division in Arabidopsis (Arabidopsis thaliana; Okazaki et al., 2015). Taken together, these findings suggest that the massive accumulation of membrane PI4P and PI(4,5)P2 resulting from terminated GH1 production could disturb organelle development and cytoarchitecture.
PI(4,5)P2 Specifically Inhibits Arp2/3 Complex-Mediated Actin Polymerization in Vitro
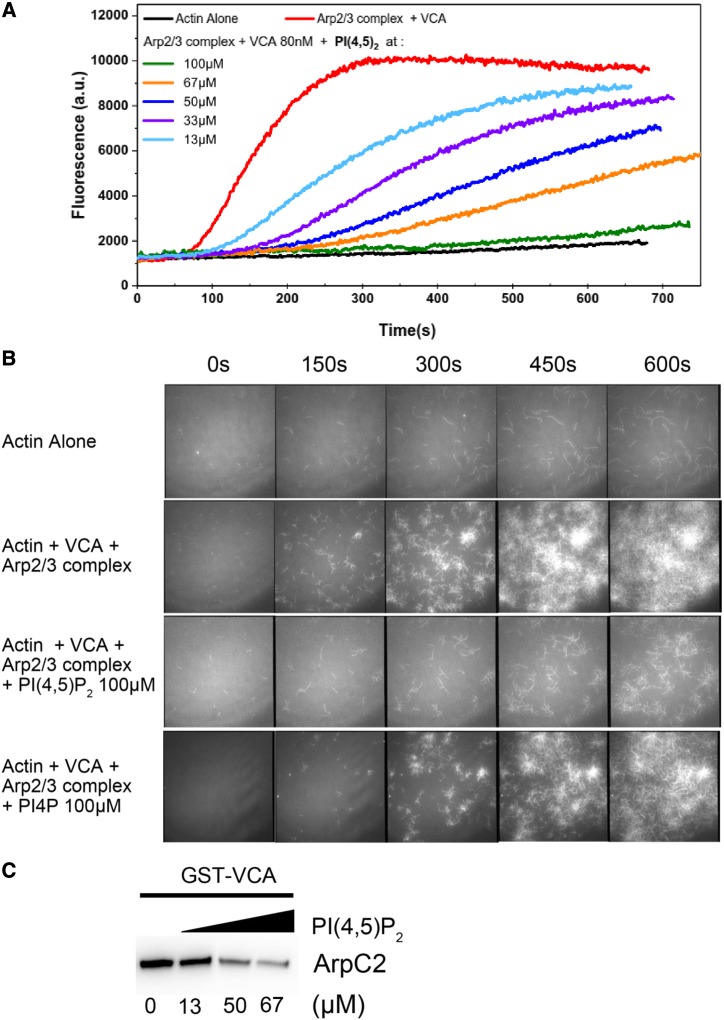
Overall, the present findings imply that maintaining proper balance between PI4P and PI(4,5)P2 levels via GH1 phosphatase is critical to cytoskeletal organization and organelle integrity. We therefore hypothesized that the collapsed organelles resulting from the overaccumulation of PI4P and PI(4,5)P2 were associated with disruption of the actin cytoskeleton. Nevertheless, the molecular mechanisms underlying PI4P- and PI(4,5)P2-induced modulation of the actin cytoskeleton remain elusive. Surprisingly, PI4P and PI(4,5)P2 were involved in the Arp2/3 complex-mediated actin polymerization in vitro (Supplemental Fig. S11). The highly conserved Arp2/3 complex functions as a nucleator of actin filament dynamics in a wide range of eukaryotic cells and consists of seven subunits: two actin-related proteins, Arp2 and Arp3, stabilized in an inactive state by five other subunits (Pollard, 2007). However, the precise function of the Arp2/3 complex in plant cells remains enigmatic, and especially little is known about the counterparts and distinction of the Arp2/3 complex in rice. To determine whether PI4P and PI(4,5)P2 are crucial lipid molecules for Arp2/3 complex action, Arp2/3 activity was assayed by spectrofluorimetry and total internal reflection fluorescence microscopy (TIRFM) with the purified porcine Arp2/3 complex, which is accepted and generally used for eukaryotic assays. The Arp2/3 complex stimulated actin polymerization 10-fold in conjunction with the indispensable VCA domain of WASP (Fig. 4A). However, both PI4P and PI(4,5)P2 inhibited Arp2/3 complex-mediated actin polymerization (Supplemental Fig. S11A), in which PI(4,5)P2 had more dramatic effect in a dose-dependent manner despite the analogous structures of PI(4,5)P2 and PI4P (Fig. 4A; Supplemental Fig. S11, A and B). These findings suggest that PI(4,5)P2 could inhibit the VCA-induced Arp2/3 complex activity. As expected, TIRFM further revealed that activation of the Arp2/3 complex via the VCA protein resulted in the generation of branched actin filaments, a hallmark of Arp2/3 complex-mediated actin filament nucleation (Fig. 4B). However, fewer actin branched junctions formed in the presence of both PI(4,5)P2 and PI4P, most notably PI(4,5)P2 (Fig. 4B; Supplemental Video S1), suggesting that PI(4,5)P2 could specifically inhibit Arp2/3 complex-mediated actin polymerization. Moreover, the elevated VCA proteins partly released the PI(4,5)P2-inhibited kinetics of actin polymerization (Supplemental Fig. S11C). Accordingly, a pull-down assay showed that PI(4,5)P2 could compete with VCA proteins binding to the Arp2/3 complex (Fig. 4C), suggesting that PI(4,5)P2 might act as a competitive inhibitor of the NPF-activated Arp2/3 complex. Overall, these findings imply that although PI(4,5)P2 was only observed in vitro to inhibit Arp2/3 complex-mediated actin polymerization, the Arp2/3 complex might function as the molecular target of PI(4,5)P2-dependent regulation.
Figure 4.
PI(4,5)P2 specifically inhibits Arp2/3 complex-mediated actin polymerization in vitro. A, Spectrofluorimetry assay using pyrene-labeled actin to monitor polymerization. PI(4,5)P2 inhibited VCA-promoted Arp2/3 complex activity in a dose-dependent manner. a.u., Arbitrary units. B, Actin polymerization monitored by TIRFM. In the presence of PI(4,5)P2, the branches initiated by the Arp2/3 complex were dramatically inhibited, whereas its structural analog, PI4P, had a much weaker inhibiting effect. C, Pull-down assay of the Arp2/3 complex with GST-VCA under elevated concentrations of PI(4,5)P2. Anti-ArpC2 was used for the immunoblotting assay. ArpC2 is a subunit of the Arp2/3 complex.
DISCUSSION
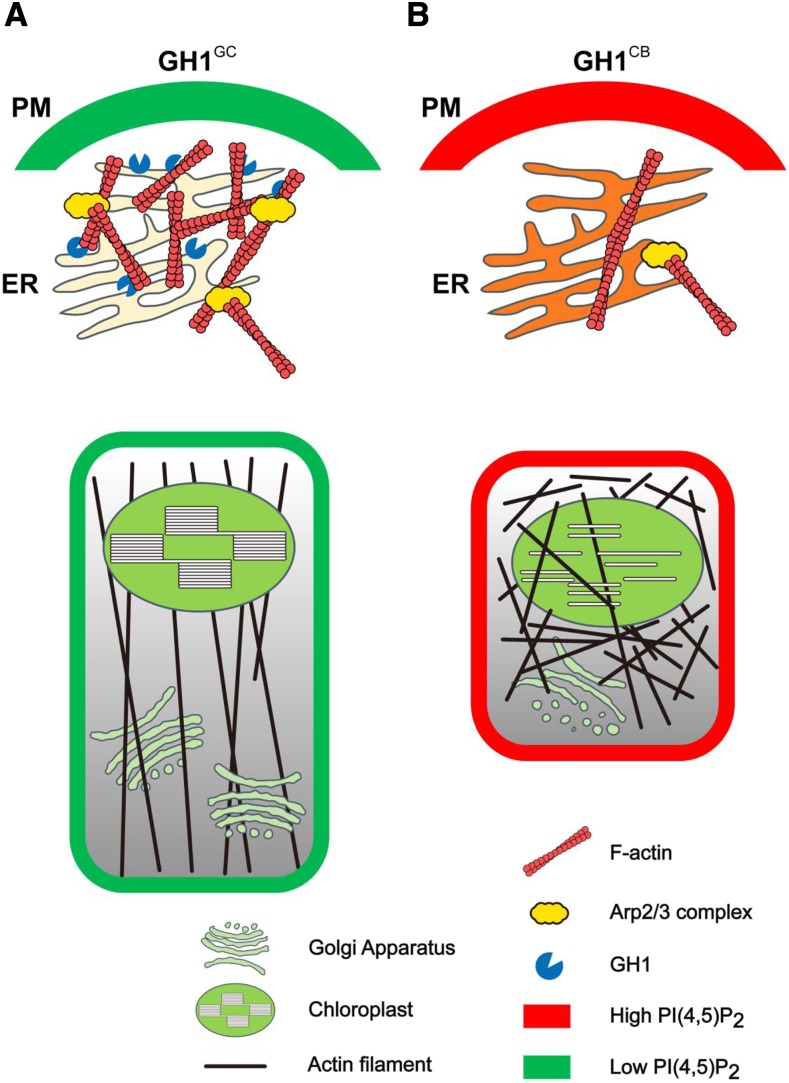
Based on our results here, we hypothesized that massive accumulation of PI(4,5)P2 could disrupt the organization of Arp2/3 complex-mediated actin polymerization, suppressing cell elongation. We subsequently proposed a working model to describe the role of GH1 phosphatase-controlled membrane PI(4,5)P2 in cell morphogenesis. Briefly, ER-localized GH1GC could specifically hydrolyze membrane PI4P and PI(4,5)P2, maintaining PI dynamics and PI(4,5)P2 distribution on the cell PM. Generally, the normal activated Arp2/3 complex initiates actin polymerization at the cell cortex under relatively low PI(4,5)P2 levels, nucleating the branched actin filament network for specific formation of the actin cytoskeleton. Therefore, these functional components enable cells to develop and elongate normally (Fig. 5A). However, the GH1CB mutation promotes endogenous PI(4,5)P2 levels in the PM and leads to dysfunctional Golgi apparatus and chloroplasts, which might suppress cell development (Fig. 5B). Here, the possible mechanism was supposed that the overaccumulation of PI(4,5)P2 resulting from the dysfunction of SAC phosphatase GH1 could inhibit Arp2/3 complex-mediated actin polymerization. Failed nucleation of the branched actin network at the cell cortex subsequently might disrupt the organization of the actin cytoskeleton and disorder cell morphogenesis. In future work, the counterparts of the Arp2/3 complex in rice should be fully identified, and revealing the relationship between PI(4,5)P2 and the Arp2/3 complex in vivo will further advance our understanding of plant growth and development.
Figure 5.
A proposed working model of the role of GH1 phosphatase-controlled membrane PI(4,5)P2 in actin cytoskeleton organization during cell morphogenesis. A, ER-localized GH1GC could specifically hydrolyze membrane PI4P and PI(4,5)P2, maintaining PI dynamics and PI(4,5)P2 distribution on the cell pm. In general, the activated Arp2/3 complex initiates actin polymerization and nucleates the branched actin filament networks for specific formation of the actin cytoskeleton. Thus, these functional components enable cells to develop and elongate normally. B, However, malfunction of GH1CB directly elevated endogenous PI(4,5)P2 levels at the PM and resulted in dysfunctional Golgi apparatus and chloroplasts, which might suppress cell development. Here, the possible mechanism was proposed that the overaccumulation of PI(4,5)P2 resulting from dysfunction of the SAC phosphatase GH1 could inhibit Arp2/3 complex-mediated actin polymerization, thereby disordering the cell morphogenesis.
Although SAC phosphatases are essential regulators of PI-signaling networks, little is known about their biochemical and cellular functions in plants. In Arabidopsis, AtSAC1 has been well characterized as a specific PI(3,5)P2 phosphatase localized on the Golgi apparatus, with truncation causing defects in cell morphogenesis and cell wall synthesis (Zhong et al., 2005). Moreover, AtSAC7 has been shown to prefer PI4P phosphatase during polarized expansion of root hair cells, thereby regulating the accumulation of PI4P in membrane compartments at the tips of growing root hairs (Thole et al., 2008). Meanwhile, an unknown subgroup of tonoplast-associated enzymes from Arabidopsis, SAC2 to SAC5, was recently found to be involved in the conversion between PI(3,5)P2 and PI3P, thereby affecting vacuolar morphology (Nováková et al., 2014). These findings suggest that members of the plant SAC phosphatase family show substrate preference and specific subcellular localization. In contrast, the results of this study provide evidence that integral ER-localized GH1 phosphatase is required for cell elongation in rice, suggesting a crucial role of SAC phosphatase in crop growth and development. Moreover, it was also observed that the GH1 phosphatase is preferential to bind to and hydrolyze PI4P and PI(4,5)P2 (Fig. 2, B–D), suggesting that GH1 specifically regulates the metabolism of membrane PI4P and PI(4,5)P2, thereby maintaining PI abundance in rice.
Dysfunction of GH1 led to massive accumulation of PI(4,5)P2 and PI4P (Fig. 2, E and F; Supplemental Fig. S4, H and I), while excessive GH1 caused their depletion (Supplemental Fig. S5, I and J), suggesting that homeostasis of PIs is pivotal for plant growth and development. In line with this, a previous study revealed that proper balance between PI4P and PI(4,5)P2 was necessary for clathrin-dependent endocytosis in the tip of pollen tubes (Zhao et al., 2010), with too much or too little impairing rapid tip growth. This supports the suggestion that flexible turnover of PIs is an important requirement for normal cell morphology. Low abundance and rapid turnover of these lipids provides a sensitive mechanism for fine-tuning protein targeting and function (Boss and Im, 2012). In plants, although activators of the Arp2/3 complex have been somewhat revealed, the determinate counterparts of the Arp2/3 complex remain largely unknown (Frank et al., 2004; Harries et al., 2005; Zhang et al., 2008). Nevertheless, the rice functional WAVE/SCAR protein TUTOU1 (TUT1) was found to promote actin nucleation and polymerization and contribute to rice development, in which the tut1 mutants showed defects in the arrangement of actin filaments and had short roots and degenerated small panicles with reduced plant height (Bai et al., 2015). This study implied that the WAVE/SCAR protein TUT1-activated Arp2/3 complex could control rice plant and panicle development via actin organization in rice, which coincides with the action of GH1 for rice morphogenesis. Moreover, in our study, NIL-GH1CB showed obvious hypersensitivity in root elongation to the inhibitor of the Arp2/3 complex, CK-548, which has been reported to insert into the hydrophobic core of Arp3 and alter its conformation (Nolen et al., 2009), over NIL-GH1GC (Supplemental Fig. S12). Therefore, we supposed that overaccumulation of PI(4,5)P2 resulting from mutated GH1 might disrupt Arp2/3 complex-mediated actin polymerization, linking PI metabolism and modulation of the WAVE/SCAR-Arp2/3 complex. These findings at least suggest that the Arp2/3 complex possibly acts as the molecular target of PI(4,5)P2-dependent modulation in eukaryotes, providing new insight into the relationship between membrane PI homeostasis and actin cytoskeleton organization. However, despite these findings, the precise molecular mechanisms underlying the action of PM PIs during activation of the Arp2/3 complex in vivo remain enigmatic. Accordingly, it is still difficult and time consuming to identify the mutants of the Arp2/3 complex components distinctly in rice, which somewhat limits our advanced understanding of the function of the plant Arp2/3 complex. Nevertheless, future analysis of the membrane PI(4,5)P2 and PI4P sensors associated with spatiotemporal colocalization of actin filaments in vivo will therefore help determine the relationship between membrane PI homeostasis and actin cytoskeleton organization for cell development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa) parental indica variety GC was crossed with japonica variety CB to generate F1 plants, which were then backcrossed with CB plants to produce BC1F1 seeds. Repetitive backcrossing and marker-assisted selection generated several plants in which the region around the GH1 locus was heterozygous, while almost all other regions were homozygous for CB, allowing segregation of populations and fine-mapping. A NIL for GH1 was subsequently developed from BC5F2 generations, containing a 50-kb chromosomal region of GC at the GH1 locus under the CB genetic background. All rice plants were cultivated in experimental fields in Shanghai and Hainan, China, under natural growth conditions. For assay of root length, the seeds were surface sterilized and germinated in one-half-strength Murashige and Skoog (MS) medium in normal conditions. The small molecule inhibitor CK-548 was diluted into medium with 10 and 30 µm.
Fine-Mapping of GH1 and Bulked Segregant Analysis
A total of 186 BC2F2 plants were used to fine-map GH1 at chromosome 2 using grain number per panicle. Subsequently, molecular markers NB-2-80 and NB-2-93 flanking GH1 were used to assess the recombinants in 6,379 BC3F2 plants. To further determine the location of GH1, markers were developed to screen plants showing homozygous recombination in the BC3F3 progeny. It was then narrowed to a region between the loci defined by markers NB-2-83c and NB-2-83d by identifying the agronomic traits of recombinant plants. Candidate GH1 genes from GC and CB were then sequenced and compared. The primers used for map-based cloning and the genotyping assays are listed in Supplemental Table S1. Bulked segregant analysis-assisted mapping of GH1 was performed as described previously (Takagi et al., 2013).
Transgene Construction and Plant Transformation
The gene-editing construct of GH1 for CRISPR/Cas9 was designed as previously described (Ma et al., 2015). To produce overexpressing transgenic plants, the full-length coding sequence of GH1 was amplified from GC and cloned into plant binary vector pCAMBIA1306 under the control of the cauliflower mosaic virus 35S promoter. Agrobacterium tumefaciens-mediated transformation of rice with strain EHA105 was then performed as described previously (Hiei et al., 1994). All constructs were confirmed by sequencing. The PCR primers are given in Supplemental Table S1.
RNA Extraction and Reverse Transcription Quantitative PCR
Total RNA was extracted from different leaves using Trizol reagent (Invitrogen). Reverse transcription was performed using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo) using 500 ng of total RNA. Reverse transcription quantitative PCR analysis was carried out using the ABI 7300 Real Time PCR System with Fast Start Universal SYBR Green Master Mix and ROX (Roche), and data were analyzed using the 2−ΔΔCT method. Rice UBQ5 gene transcripts were used as a reference. The relevant primers for gene amplification are given in Supplemental Table S1.
Subcellular Localization of GH1
To determine the subcellular localization of GH1, the GH1 fragment was inserted into the pA7-GFP plasmid and transformed into rice protoplasts. Organelle marker CD3-959-mCherry was used as the ER marker (Nelson et al., 2007). GFP (excitation, 488 nm; emission, 507 nm) and mCherry (excitation, 587 nm; emission, 610 nm) fluorescence in rice protoplasts was detected using an LSM 880 confocal laser-scanning microscope (Zeiss). Sequences of the PCR primers used for vector construction are given in Supplemental Table S1.
Transmission Electron Microscopy
For scanning transmission electron microscopy of Golgi apparatus and chloroplast observation, leaves from 10-d-old rice seedlings grown on one-half-strength MS plates were fixed in situ. Briefly, the leaves were removed and transferred immediately to vials with fresh primary fixative (1% [v/v] paraformaldehyde, 2.5% [v/v] glutaraldehyde, and phosphate buffer, pH 7.2) followed by 4 h of incubation at room temperature. The samples were then rinsed in phosphate buffer and fixed in 1% (v/v) osmium tetroxide overnight at 4°C before dehydration in an ascending alcohol series. After fixing, the samples were stepwise infiltrated with epoxy resin and then embedded in Epon-812 resin before slicing into thin sections using a diamond knife. The samples were then placed on nickel grids and double stained with 2% (v/v) aqueous uranyl acetate and lead citrate for observation using a Hitachi H-7650 transmission electron microscope. By use of different views from diverse plants, the layers of Golgi apparatus and thylakoids were calculated in different cut sections, which were layered with discal structure and easily discernible in different images. For x-ray imaging, the rice culms were fixed in 50% (v/v) ethanol, 5% (v/v) glacial acetic acid, and 5% (v/v) formaldehyde overnight at 4°C and then dehydrated in a graded alcohol series before critical point drying. The prepared samples were then examined using an Xradia 520 Versa x-ray imager (Zeiss) and then numbered using ImageJ software. For the semithin sectioning of roots, the experiment was performed as described previously (Guo et al., 2018).
Protein Expression and Phosphatase Activity Assay
The highly efficient Bac-to-Bac baculovirus expression system (Invitrogen) was used for protein expression in Sf9 insect cells. The coding sequences of GH1 and its mutated version were inserted into the pFastBacHT-C vector. Culturing of the insect cells and generation of the recombinant bacmid and recombinant baculovirus were performed according to the manufacturer’s handbook. A Mem-PER Plus Membrane Protein Extraction Kit and Ni-NTA resin (Thermo Scientific) were then used to purify the proteins following the manufacturer’s protocol. To assay specific lipid phosphatase activity in vitro, a Malachite Green Assay Kit (Echelon Biosciences) was used to detect the liberated phosphate among different PI substrates according to the manufacturer’s protocol. The primers used for vector construction are listed in Supplemental Table S1.
In Vitro Lipid-Binding Assay
For lipid-binding assay in vitro, the PIP strips (Echelon Biosciences) were blocked in a solution of 5% (w/v) fatty acid-free BSA in Tris-buffered saline plus Tween 20 for 1 h at room temperature, and then the blocked membrane lipid strips were incubated with 0.05 mg mL−1 fusion proteins for 3 h with gentle agitation at room temperature. The membrane strips were then washed extensively with wash buffer three times. The fusion proteins were detected with horseradish peroxidase-conjugated mouse anti-His monoclonal antibody (ABclonal, AE028; 1:3,000 dilution) and visualized by enhanced chemiluminescence.
In Vivo Measurements of PI4P and PI(4,5)P2 Levels Using ELISA
The PIs were extracted from rice as described previously (Drøbak et al., 2000), using 50 mg of fresh leaves for each sample. Measurements of PI4P and PI(4,5)P2 levels in the different samples were then performed using a PI(4)P and PI(4,5)P2 Mass ELISA Assay Kit (Echelon Biosciences) by means of competitive ELISA. Based on the assay protocol, the colorimetric signals were read at an absorbance of 450 nm and were inversely proportional to the amount of PI4P and PI(4,5)P2 extracted from the cells.
Staining and Visualization of Actin Filaments
To visualize the actin cytoskeleton in the rice cells, root tips from 5-d-old rice seedlings grown on one-half-strength MS plates were immersed in PEM buffer (100 mm PIPES, 10 mm EGTA, 5 mm MgSO4, and 0.3 m mannitol, pH 6.9) with 2% (v/v) glycerol. ActinGreen488 Reagent (Thermo Scientific) was added directly to the buffer to stain the actin filaments followed by incubation at room temperature for 35 min. The green fluorescence-labeled actin filaments (excitation, 488 nm; emission, 507 nm) were then viewed using an LSM 880 confocal laser-scanning microscope (Zeiss). More than 10 views for each allele and at least one to two cells in every view were observed. To characterize the angular orientation distribution of actin filaments, the fully automated and optimized two-dimensional Discrete Fourier Transform method was applied on the samples from each confocal image as described in a previous study (Marquez, 2006). Simply, the custom-written script running under MATLAB was generously provided by Keith Carney (University of Utah). The RGB (Red, Green, Blue) confocal images were changed to eight-bit black-and-white TIFF images in Photoshop and used as subsamples. To further minimize errors, the subsamples were normalized by adjusting their orientation relative to the cell longitudinal axis. In each allele, 10 representative and different subsamples containing at least 20 cells were analyzed and fitted. The radar chart based on the MATLAB programming language was used to statistically analyze and fit the direction of the actin filaments (360°). The most representative radar chart from one cell in each allele is shown. For comparing the length and number of actin filaments, the consecutive line-like and obviously stained actin filaments along the longitudinal axis from one side to the other in each view were numbered as the assumed length of actin filaments, and the specific line-like fluorescent signals were calculated as the assumed number of actin filaments by use of ImageJ software.
In Vitro Actin Polymerization Assay
To determine the effects of PI4P and PI(4,5)P2 on Arp2/3 complex-mediated actin-nucleating activity in vitro, pyrene-labeled actin assays and monitoring of the branching reaction were performed as described previously (Higgs et al., 1999; Zhang et al., 2010). The purified Arp2/3 protein complex from porcine brain (Cytoskeleton) and GST-tagged VCA domain of human (Homo sapiens) WASP protein (Cytoskeleton) were used to initiate actin nucleation. Monomeric actin (10% [v/v] pyrene labeled) was incubated with the Arp2/3 complex and GST-VCA protein in the presence or absence of PI4P and PI(4,5)P2 at the indicated concentrations in buffer G (2 mm Tris-HCl, pH 8, 0.01% [w/v] NaN3, 0.2 mm CaCl2, 0.2 mm ATP, and 0.2 mm DTT) at room temperature. The time course of actin polymerization was then tracked by monitoring the changes in pyrene fluorescence using a QuantaMaster Luminescence QM 3 PH Fluorometer (Photon Technology International) with excitation set at 365 nm and emission at 407 nm immediately after adding 1× KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, and 10 mm imidazole-HCl, pH 7).
Visualization of Actin Assembly Using TIRFM
Direct visualization of actin assembly was carried out using TIRFM as previously described (Amann and Pollard, 2001; Shi et al., 2013). Briefly, the flow cells were preincubated with 25 nm NEM-myosin and equilibrated with 1% (w/v) bovine serum albumin, then washed with fluorescence buffer (10 mm imidazole, pH 7, 50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 50 mm DTT, 0.2 mm ATP, 50 µm CaCl2, 15 mm Glc, 20 µg mL−1 catalase, 100 µg mL−1 Glc oxidase, and 0.5% [w/v] methylcellulose). Preassembled actin filaments (25 nm) in the fluorescence buffer were then injected into the flow cells and allowed to settle. To assess the effect of PIs on actin branching, PI4P and PI(4,5)P2 from porcine brain (Avanti) were perfused at various concentrations into the flow cells. The actin filaments were then visualized using an Olympus IX-71 microscope equipped with a 60×, 1.45-numerical aperture Planapo objective (Olympus) by TIRFM illumination. Time-lapse imaging of the actin filaments was captured at 3-s intervals.
GST Pull Down
The VCA domain of human WASP protein was expressed in a bacterial expression system as a GST-tagged fusion protein (Cytoskeleton). The seven protein subunits of the Arp2/3 protein complex were purified from bovine brain (Cytoskeleton). The GST-VCA fusion protein associated with the glutathione-agarose 4B (GE Healthcare) was incubated with the Arp2/3 complex in pull-down buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, 0.5% [v/v] Triton X-100, and 5% [v/v] glycerol) for 2 h at 4°C. Various concentrations of PI(4,5)P2 from porcine brain (Avanti) were used for competition. The GST beads were washed and eluted for immunoblot assays using anti-ArpC2 antibody (Abcam, ab96779; 1:3,000 dilution).
Accession Numbers
The gene sequence from this article can be found in the Rice Annotation Project Database or Rice Genome Annotation Project under the following accession numbers: Os02g0554300 or LOC_Os02g34884 (GH1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genetic identification of GH1.
Supplemental Figure S2. Bulked segregant analysis-assisted mapping of GH1.
Supplemental Figure S3. Genotyping identification of the mutants edited by CRISPR/Cas9.
Supplemental Figure S4. Deletion of GH1 suppresses rice growth and development.
Supplemental Figure S5. Overexpression of GH1 had a weaker effect on plant and panicle morphogenesis in rice.
Supplemental Figure S6. Phylogenetic analysis and amino acid sequence alignment of GH1 homologs from different species.
Supplemental Figure S7. GH1 is a membrane-localized protein.
Supplemental Figure S8. Overaccumulation of PI(4,5)P2 in roots could disorder actin organization.
Supplemental Figure S9. Mutation of GH1 suppressed root development in rice.
Supplemental Figure S10. Mutation of GH1 disrupted the structural integrity of Golgi apparatus and chloroplasts.
Supplemental Figure S11. Inhibitory action of PI(4,5)P2 toward the Arp2/3 complex is specific and is released by elevated amounts of VCA.
Supplemental Figure S12. NIL-GH1CB showed hypersensitivity to the inhibitor of the Arp2/3 complex.
Supplemental Table S1. Primers used in this study.
Supplemental Video S1. Visualization of the Arp2/3 complex-nucleated actin-branching networks using TIRFM.
Acknowledgments
We thank Min Shi (Institute of Plant Physiology and Ecology, Shanghai Institute for Biological Sciences, Chinese Academic of Sciences) for technical support with the transgenic assay. We thank Xiaoyan Gao, Zhiping Zhang, and Jiqin Li (Institute of Plant Physiology and Ecology, Shanghai Institute for Biological Sciences, Chinese Academic of Sciences) for technical support. We thank Chunjie Cao (Carl Zeiss) for x-ray microscopy observation.
Footnotes
This work was supported by the Ministry of Science and Technology of the People’s Republic of China (grant no. 2016YFD0100902), the National Natural Science Foundation of China (grant no. 31788103), the Chinese Academy of Sciences (grant nos. QYZDY-SSW-SMC023 and XDB27010104), the China Postdoctoral Science Foundation (grant no. 2018M642102), the Science and Technology Commission of Shanghai Municipality (grant no. 18JC1415000), the CAS-Croucher Funding Scheme for Joint Laboratories, and the National Key Laboratory of Plant Molecular Genetics.
References
- Amann KJ, Pollard TD (2001) Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci USA 98: 15009–15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Zhu X, Wang Q, Zhang J, Chen H, Dong G, Zhu L, Zheng H, Xie Q, Nian J, et al. (2015) Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organization and panicle development. Plant Physiol 169: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. (2013) Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol Rev 93: 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss WF, Im YJ (2012) Phosphoinositide signaling. Annu Rev Plant Biol 63: 409–429 [DOI] [PubMed] [Google Scholar]
- Bothe I, Deng S, Baylies M (2014) PI(4,5)P2 regulates myoblast fusion through Arp2/3 regulator localization at the fusion site. Development 141: 2289–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice SE, Alford CW, Cowart LA (2009) Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem 284: 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P (2015) PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak BK, Brewin NJ, Hernandez LE (2000) Extraction, separation, and analysis of plant phosphoinositides and complex glycolipids. Methods Mol Biol 141: 157–174 [DOI] [PubMed] [Google Scholar]
- Ercetin ME, Gillaspy GE (2004) Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol 135: 938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD (2001) Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell 12: 2396–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Egile C, Dyachok J, Djakovic S, Nolasco M, Li R, Smith LG (2004) Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc Natl Acad Sci USA 101: 16379–16384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth K, Lin F, Menzel W, Krishnamoorthy P, Stenzel I, Heilmann M, Heilmann I (2017) Guilt by association: A phenotype-based view of the plant phosphoinositide network. Annu Rev Plant Biol 68: 349–374 [DOI] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong NQ, Shi CL, Ye WW, Gao JP, Shan JX, Lin HX (2018) GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30: 871–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries PA, Pan A, Quatrano RS (2005) Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I. (2016) Phosphoinositide signaling in plant development. Development 143: 2044–2055 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD (1999) Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38: 15212–15222 [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Menon AK (2014) Lipid landscapes and pipelines in membrane homeostasis. Nature 510: 48–57 [DOI] [PubMed] [Google Scholar]
- Hsu F, Mao Y (2013) The Sac domain-containing phosphoinositide phosphatases: Structure, function, and disease. Front Biol (Beijing) 8: 395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkowska A, Niewczas I, Anderson KE, Durrant TN, Clark J, Stephens LR, Hawkins PT (2014) A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv Biol Regul 54: 131–141 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim S, Nahm M, Kim E, Kim TI, Yoon JH, Lee S (2011) The phosphoinositide phosphatase Sac1 is required for midline axon guidance. Mol Cells 32: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Blanchoin L, Staiger CJ (2015) Signaling to actin stochastic dynamics. Annu Rev Plant Biol 66: 415–440 [DOI] [PubMed] [Google Scholar]
- Liu Y, Boukhelifa M, Tribble E, Morin-Kensicki E, Uetrecht A, Bear JE, Bankaitis VA (2008) The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol Biol Cell 19: 3080–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Manford A, Xia T, Saxena AK, Stefan C, Hu F, Emr SD, Mao Y (2010) Crystal structure of the yeast Sac1: Implications for its phosphoinositide phosphatase function. EMBO J 29: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez JP. (2006) Fourier analysis and automated measurement of cell and fiber angular orientation distributions. Int J Solids Struct 43: 6413–6423 [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B (2013) A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155: 830–843 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD (2009) Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460: 1031–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková P, Hirsch S, Feraru E, Tejos R, van Wijk R, Viaene T, Heilmann M, Lerche J, De Rycke R, Feraru MI, et al. (2014) SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc Natl Acad Sci USA 111: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Miyagishima SY, Wada H (2015) Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 27: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK (2010) Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem 79: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskot R, Pejchar P, Staiger CJ, Potocký M (2014) When fat is not bad: The regulation of actin dynamics by phospholipid signaling molecules. Front Plant Sci 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. (2007) Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct 36: 451–477 [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE (2013) New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 14: 7–12 [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Lappalainen P (2010) Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev 90: 259–289 [DOI] [PubMed] [Google Scholar]
- Shi M, Xie Y, Zheng Y, Wang J, Su Y, Yang Q, Huang S (2013) Oryza sativa actin-interacting protein 1 is required for rice growth by promoting actin turnover. Plant J 73: 747–760 [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144: 389–401 [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Emr SD (2013) ER-PM connections: Sites of information transfer and inter-organelle communication. Curr Opin Cell Biol 25: 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, et al. (2013) QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74: 174–183 [DOI] [PubMed] [Google Scholar]
- Takasuga S, Sasaki T (2013) Phosphatidylinositol-3,5-bisphosphate: Metabolism and physiological functions. J Biochem 154: 211–218 [DOI] [PubMed] [Google Scholar]
- Thole JM, Vermeer JE, Zhang Y, Gadella TW Jr., Nielsen E (2008) Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Hussey PJ (2017) Connecting membranes to the actin cytoskeleton. Curr Opin Plant Biol 40: 71–76 [DOI] [PubMed] [Google Scholar]
- Wei HC, Sanny J, Shu H, Baillie DL, Brill JA, Price JV, Harden N (2003) The Sac1 lipid phosphatase regulates cell shape change and the JNK cascade during dorsal closure in Drosophila. Curr Biol 13: 1882–1887 [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Zhang C, Szymanski DB (2013) ARP2/3-dependent growth in the plant kingdom: SCARs for life. Front Plant Sci 4: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Mallery EL, Schlueter J, Huang S, Fan Y, Brankle S, Staiger CJ, Szymanski DB (2008) Arabidopsis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20: 995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qu X, Bao C, Khurana P, Wang Q, Xie Y, Zheng Y, Chen N, Blanchoin L, Staiger CJ, et al. (2010) Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Mao YS, Janmey PA, Yin HL (2012) Phosphatidylinositol 4,5 bisphosphate and the actin cytoskeleton. Subcell Biochem 59: 177–215 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yan A, Feijó JA, Furutani M, Takenawa T, Hwang I, Fu Y, Yang Z (2010) Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell 22: 4031–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Nairn CJ, Wood-Jones A, Morrison WH III, Ye ZH (2005) Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17: 1449–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2003) The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol 132: 544–555 [DOI] [PMC free article] [PubMed] [Google Scholar]