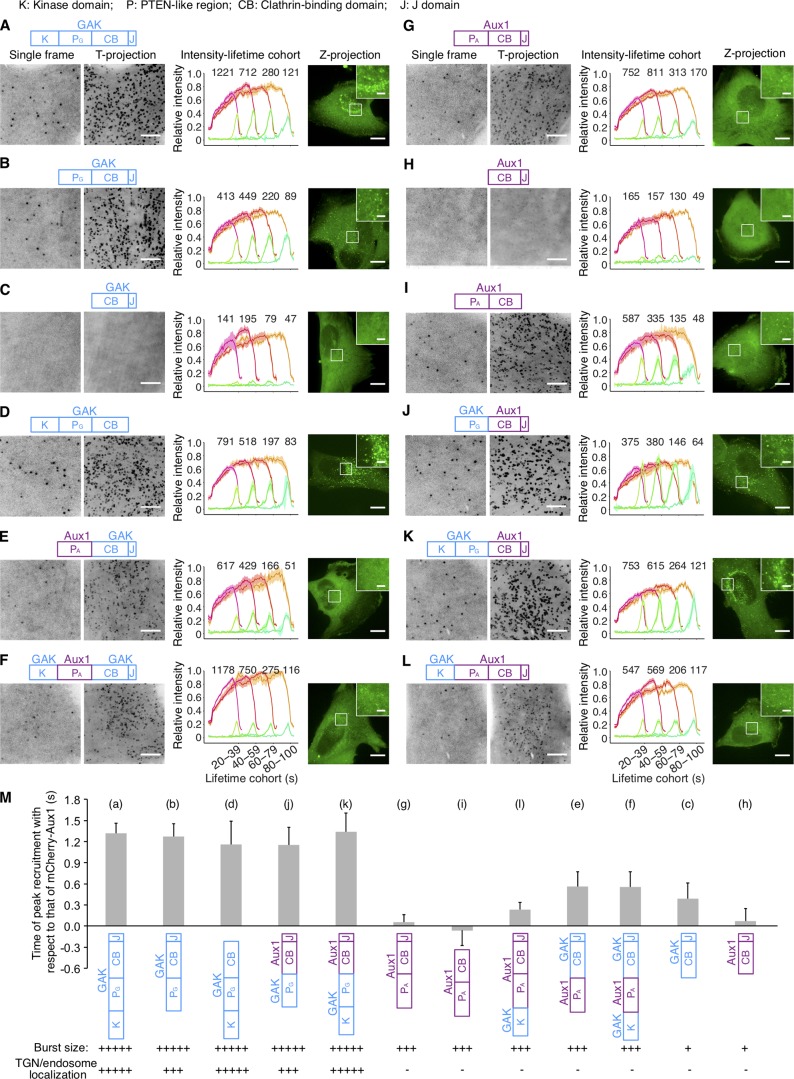
Figure 4.
Influence of PTEN-like domains on Aux1 and GAK recruitment. (A–L) Bottom surfaces of gene-edited CLTA-TagRFP+/+ cells transiently expressing indicated EGFP-tagged constructs of Aux1 or GAK, imaged by TIRF microscopy every 1 s for 300 s. Each panel shows a schematic representation of the construct domain organization: K, kinase domain of GAK (amino acid residues 1–347); PA or PG, PTEN-like domain of GAK (360–766) or Aux1 (1–419); CB, clathrin-binding domain of GAK (767–1,222) or Aux1 (420–814); J, J domain of GAK (1,223–1,311) or Aux1 (815–910); a representative single frame and the corresponding maximum-intensity time projection of the time series as a function of time (T-projection). The plots show averaged fluorescence intensity traces (mean ± SE) of CLTA-TagRFP (red) and EGFP-fused constructs (green), from 6–13 cells per condition, grouped by cohorts according to lifetimes. The numbers of analyzed traces are shown above each cohort. The cells were also imaged in 3D by spinning-disk confocal microscopy; the images at the right of each panel show representative maximum-intensity z projections (Z-projection) from 34 sequential optical sections spaced at 0.3 µm (scale bars, 10 µm) and corresponding enlarged regions (scale bars, 2 µm). (M) Mean interval between the fluorescence maxima for the indicated EGFP-fused construct (diagram immediately below plot) and the mCherry-Aux1 burst (mean ± SD, n = 8–14 cells), in cells imaged at 0.5-s intervals for 60 s by TIRF microscopy. Below the construct schematics are qualitative estimates of the relative maximum amplitudes of fluorescence for the bursts at the plasma membrane and in regions of the TGN/endosome.