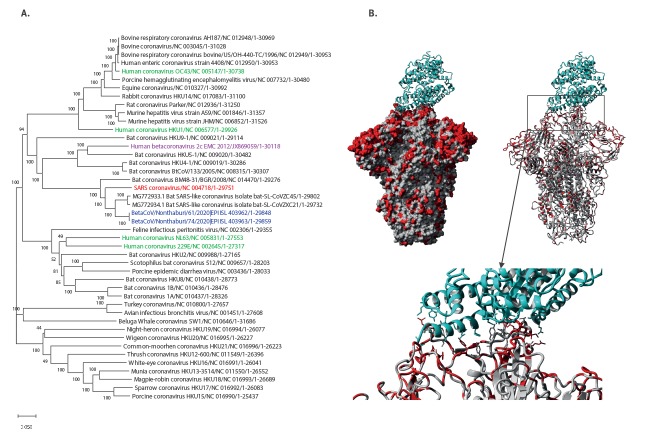
Figure 1.
Phylogenetic trees of Thai sequences in context of all coronavirus families (A) and structural mapping of mutations in the spike glycoprotein between SARS CoV (PDB:6CG [12]) and the current SARS-CoV-2 using YASARA [20] (B)
CoV: coronavirus; MERS: Middle East respiratory syndrome coronavirus; SARS: severe acute respiratory syndrome.
Panel A: blue: SARS-CoV-2; red: SARS; purple: MERS; green: common cold.
Panel B: cyan: ACE2 human host receptor; grey: CoV spike glycoprotein trimer (PDB:6ACG); red: mutations between SARS-CoV vs SARS-CoV-2.
The phylogenetic tree was created from whole genome alignment with MAFFT using the neighbour-joining method with maximum composite likelihood (MCL) model, uniform site rates and 500 bootstrap tests using MEGA X.