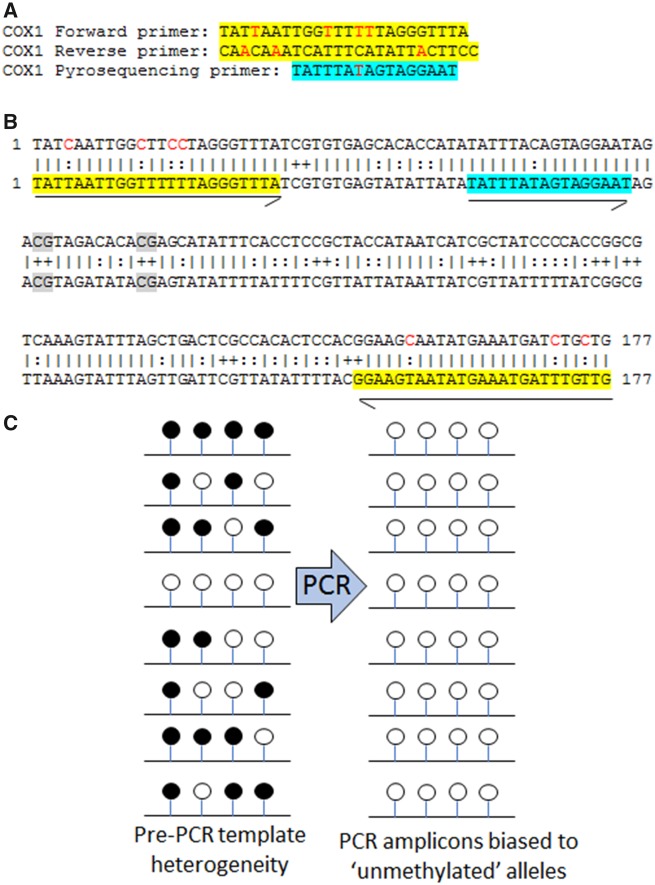
Figure 1:
PCR from bisulphite-converted DNA in regions with non-CpG methylation is biased towards amplification of unmethylated alleles if the primer contains cytosines. (A) Typical primers used for bisulphite sequencing with bases that hybridize to converted non-CpG cytosines (red text). (B) Alignment of unconverted (top) and converted (bottom) sequences with non-CpG cytosines indicated with colons and CpGs with plus signs. Locations of PCR and pyrosequencing primers highlighted in yellow and blue, respectively, and indicated with arrows. Converted non-CpG cytosines in primers in red text. (C) Schematic showing selective amplification in PCR from unmethylated alleles in regions with a high frequency of non-CpG methylation. Methylated/unconverted and unmethylated/converted cytosines indicated with black and white lollipops, respectively