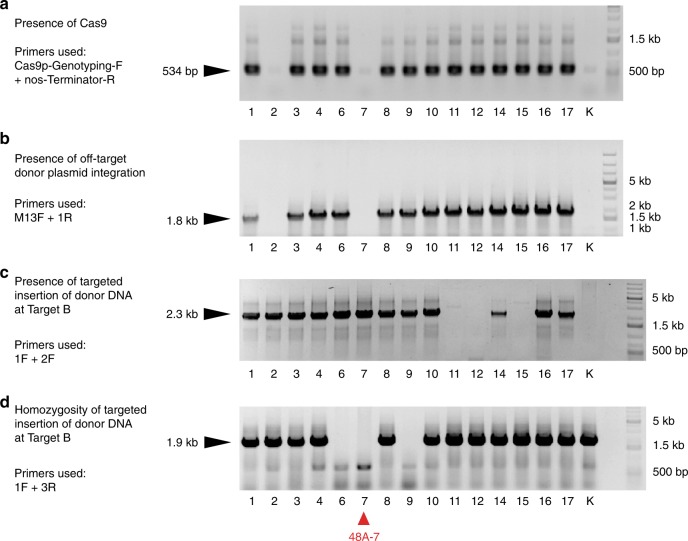
Fig. 3. Genetic segregation of the progeny of T0-48A.
Genotyping the T1 progeny of T0-48A. The purpose of each PCR experiment and the genotyping primers used are shown to the left for each gel panel. a Primers Cas9p-Genotyping-F and nos-Terminator-R amplify a 534 bp DNA fragment in plants with the Cas9 module. b Primers M13F and 1R amplify a 1.8 kb DNA fragment in plants with the off-target insertion of the pAcc-B donor plasmid. c Primers 1F and 2F amplify a 2.3 kb DNA fragment in plants with the carotenoid cassette inserted at Target B. d Primers 1F and 3R amplify a 1.9 kb genomic DNA fragment in plants unless the carotenoid cassette at Target B is homozygous. The positions of the primers used are illustrated in Figs. 1a, b, 2a. Kitaake (K) was used as the negative control. The red triangle at the bottom marks 48A-7, which is free of the co-integrated CRISPR and donor plasmids. Source data are provided as a Source Data file.