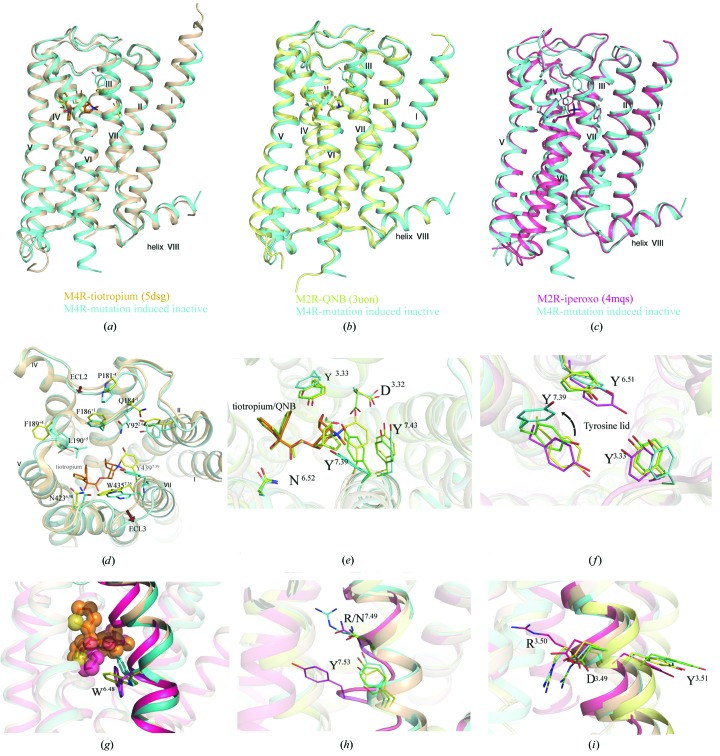
Figure 2.
Comparison of the mutation-induced inactive M4 structure with structures of M2 and M4 in inactive and active states. (a, b, c) M2 and M4 are aligned with the mutation-induced inactive M4 structure (teal blue). The M4–tiotropium structure is shown as an orange cartoon and the inactive and active M2 structures are shown as pale yellow and magenta cartoons, respectively. (d) Extracellular region comparison of mutation-induced inactive M4 and M4–tiotropium (PDB entry 5dsg) structures. (e) The highly conserved residues in orthosteric binding pockets for tiotropium and QNB. (f) The ‘tyrosine lid’ is formed by three tyrosines: Y6.51, Y3.33 and Y7.39. There is a 110° rotation of Y7.39 compared with that in M4–tiotropium; the arrow shows the rotation of Y4397.39 in the mutation-induced inactive M4 structure. (g) The different rotations of W6.48 in the mutation-induced inactive M4, active M2 and M4–tiotropium structures. (h, i) Comparison of the NPxxY and D(E)RY motifs between the inactive and active mAchR structures.