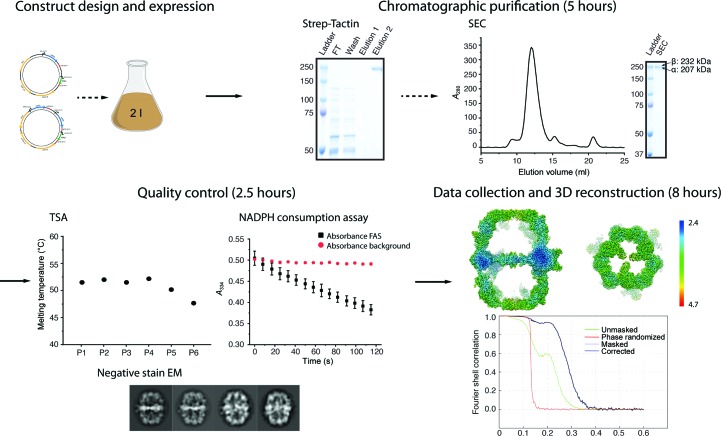
Figure 1.
Structural analysis of yeast FAS. Yeast FAS was expressed overnight from pRS vector-encoded FAS1 and FAS2 genes. Gravity flow of the cleared lysate over a Strep-Tactin column and subsequent size-exclusion chromatography (SEC) delivered pure protein within 5 h. Protein quality was monitored by NADPH consumption, thermal shift assays (TSA) and negative-stain transmission EM within 2.5 h. Thermal stability was tested for a set of conditions (P1, 100 mM sodium phosphate pH 6.5; P2, 100 mM sodium phosphate pH 7.4; P3, 100 mM sodium phosphate pH 8; P4, 100 mM sodium phosphate, 100 mM NaCl pH 7.4; P5, 100 mM Tris–HCl pH 7.4; P6, distilled water). The activity of the preparation was 2310 ± 48 mU mg−1 and the error in the melting point varied by less than 0.5°C; both values were within technical replication. Protein integrity was assessed further by negative-stain EM and 2D single-particle image analysis (within 1.5 h). CryoEM images were collected in movie mode in 4.5 h. 20 000 particles were picked automatically, of which 15 000 were selected by 2D and 3D classification, to yield a map at 3.1 Å resolution in 3.5 h of image processing.