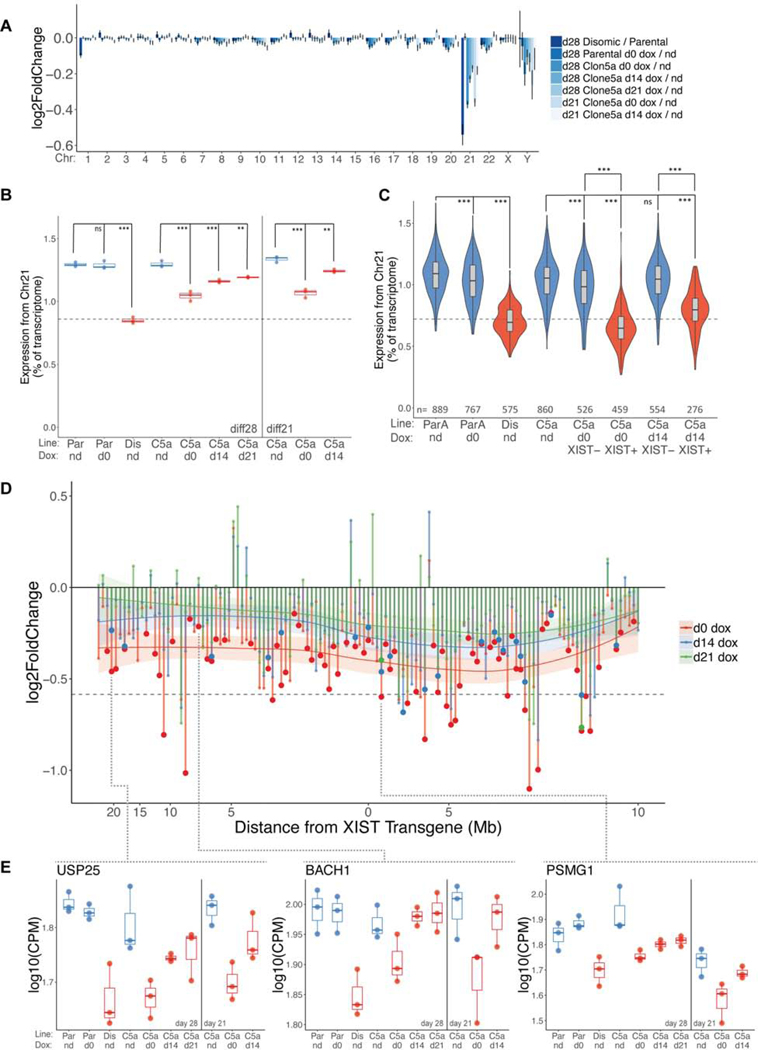
Figure 3:
Transcriptional chromosome 21 repression at all time points of XIST initiation. a) Bar graph of bulk RNAseq data of mean log2 fold change at diff28 for all detected genes on each chromosome for each 3 vs. 3 comparison. Data are represented as mean ± SEM. Note: Y chromosome data represents only 9 detected genes. b) diff21 and diff28 bulk RNAseq data. Fraction of normalized chr21 reads over all reads for each sample. n=3 independent differentiations for each condition. Dashed line represents a 33% drop in chr21 transcription from mean parental level. Samples colored in blue are trisomic; samples colored in red are disomic or functionally disomic (XIST+). c) Violin plots of scRNAseq data at diff28 showing median (horizontal line), interquartile range (rectangular box), 95% confidence interval, and the kernel probability density at each value. For each cell, the fraction of UMIs from chr21 is divided by total UMIs to determine expression from chr21. Number of cells in each sample is provided. d) Bulk RNAseq fold change between indicated comparisons for all chr21 genes with FPKM>1 and normalized average read count >10 plotted against ranked chromosomal position. Approximate chromosomal distance in megabases (Mb) relative to the XIST transgene locus is indicated on X-axis. Local average (LOESS) for each comparison is indicated by solid horizontal curves with 95% confidence intervals. Dotted horizontal line indicates 1.5-fold change. Significant differential expression (FDR ≤ 0.1) is indicated by larger dots. e) Dot plots of all samples for 3 selected chr21 genes demonstrating various silencing kinetics. Dotted lines indicate each gene’s position in (d). **p ≤ 0.01, ***p ≤ 0.001; one-way ANOVA followed by Tukey’s multiple comparisons test. Par, parental trisomic line; ParA, parental subclone A; Dis, disomic; C5A, Transgenic Clone 5a; nd, no dox; d0, day 0 dox initiation; d14, day 14 dox initiation; d21, day 21 dox initiation.