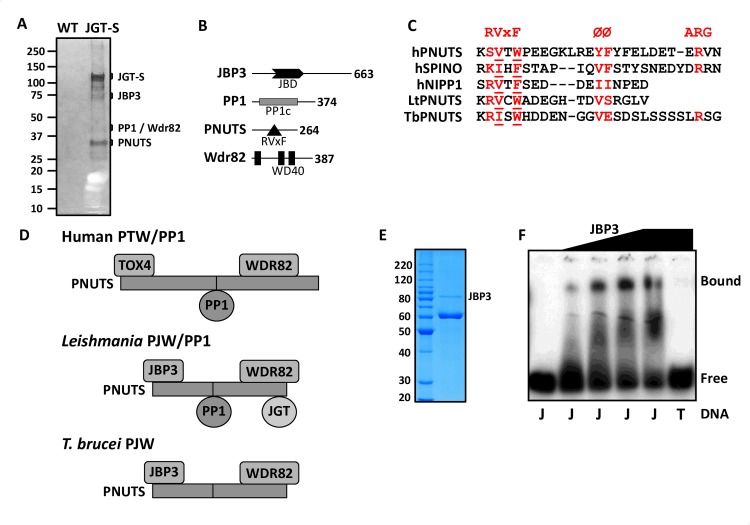
Fig 1. Identification of a novel phosphatase complex in Leishmania tarentolae.
(A) Proteins recovered from tandem affinity purification from wild-type L. tarentolae extracts (WT) and from cells expressing the strep-tagged version of JGT (JGT-S) in a single JGT KO background, were analyzed on SDS-PAGE gel and silver stained. The location of the tagged JGT (confirmed by western blot) and associated proteins are indicated based upon MS analysis of the gel fragments and their predicted molecular weight. The inherent disorder in PNUTS may explain why it migrates slower in SDS-PAGE than predicted. The additional bands in the SDS-PAGE presumably represent degradation (primarily of JGT) as indicated by the MS analysis. (B) Summary of the PJW/PP1 complex. The domain structure of each component in the complex is schematically shown. JBD, J-DNA binding domain; RVxF motif, PP1 docking motif; PP1c, catalytic domain of protein phosphatase 1; WD40, WD40 repeat. The number of amino acids in each component is indicated. (C) Structure-based sequence alignment of the PNUTS, spinophilin PP1 and NIPP1 interactive domains in humans is compared with LtPNUTS (residues 95–112) and TbPNUTS (residues 139–164), where x is any amino acid and Ø represents a hydrophobic amino acid. Critical residues in the RVxF motif are underlined. (D) Models for the PJW/PP1 complex in Leishmania and T. brucei. The models are based on the human PTW/PP1 complex where PNUTS acts as scaffold and the DNA binding protein and Wdr82 bind to the N- and C-terminus, respectively, and PP1 binds via the RVxF PP1 interaction motif (indicated by the line). PP1 is presumably not stably associated with the complex in T. brucei (see discussion) and is therefore labeled as the PJW complex. (E) Purification of recombinant LtJBP3. His-tagged rJBP3 expressed in E. coli was purified by metal affinity and size exclusion chromatography and analyzed by SDS-PAGE/Coomassie staining. The major copurified protein is the E. coli molecular chaperone GroEL. The migration of protein molecular mass standards (in kDa) is shown on the left. (F) Gel shift assays for modified and unmodified DNA substrates interacting with JBP3. 0.3 pmol radiolabeled J-DNA (J) was incubated with 0, 0.2, 0.38, 0.57 and 1 pmol of JBP3 and 0.3 pmol radiolabeled unmodified DNA (T) was incubated with 1 pmol of JBP3.