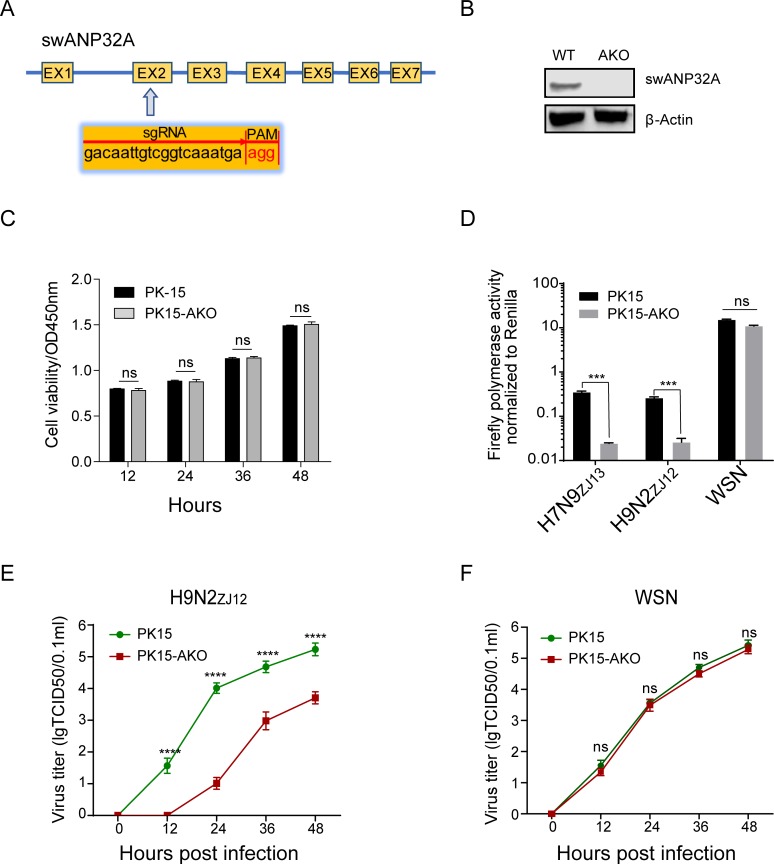
Fig 3. Knockout of ANP32A in pig cells sharply reduced avian influenza viral RNA replication.
(A) The scheme of sgRNA used for knocking out the swANP32A gene. PK15 cells were transfected with pMJ920 vector (plasmid expressing eGFP and Cas9) and gRNAs to generate swANP32A knockout cells (PK15-AKO). (B) The endogenous proteins in wild type and knockout PK15 cells were identified by western blotting using antibodies against β-actin (sc-47778, Santa Cruz) and swANP32A (Rabbit Anti-PHAP1 polyclonal antibody, bs-6083R, Bioss). (C) Wildtype and AKO PK-15 cells were seeded in 96-well plate at 1x10^4 cells per well, 10 μL of CCK-8 reagents (meilunbio, MB4420) were added at the indicated times, and then the 96-well plate was incubated at 37°C for 1.5 h. The optical density (OD) values were measured by a microplate reader at OD450nm wavelength. (D) Wild type and knockout PK15 cells were transfected with Firefly minigenome reporter, Renilla expression control, together with polymerases from the avian influenza viruses H7N9ZJ13, or H9N2ZJ12, or the human influenza virus WSN. Luciferase activity was measured at 24 h post transfection. Data are Firefly activity normalized to Renilla. Statistical differences between cells are labeled according to a one-way ANOVA followed by a Dunnett’s test; NS = not significant, ***P < 0.001. (E and F) Wild type and knockout PK15 cells were infected with avian influenza virus H9N2ZJ12 and human influenza virus WSN at an MOI of 0.1, respectively. The supernatants were sampled at 0, 12, 24, 36, 48 h post infection and the virus titers were determined by means of endpoint titration in PK15 cells. (Statistical difference between groups were labeled, according to a one-way ANOVA followed by a Dunnett’s test; NS = not significant, ****P < 0.0001).