Abstract
Enzymatic substrate promiscuity is more ubiquitous than previously thought, with significant consequences for understanding metabolism and its application to biocatalysis. This realization has given rise to the need for efficient characterization of enzyme promiscuity. Enzyme promiscuity is currently characterized with a limited number of human-selected compounds that may not be representative of the enzyme’s versatility. While testing large numbers of compounds may be impractical, computational approaches can exploit existing data to determine the most informative substrates to test next, thereby more thoroughly exploring an enzyme’s versatility. To demonstrate this, we used existing studies and tested compounds for four different enzymes, developed support vector machine (SVM) models using these datasets, and selected additional compounds for experiments using an active learning approach. SVMs trained on a chemically diverse set of compounds were discovered to achieve maximum accuracies of ~80% using ~33% fewer compounds than datasets based on all compounds tested in existing studies. Active learning-selected compounds for testing resolved apparent conflicts in the existing training data, while adding diversity to the dataset. The application of these algorithms to wide arrays of metabolic enzymes would result in a library of SVMs that can predict high-probability promiscuous enzymatic reactions and could prove a valuable resource for the design of novel metabolic pathways.
Keywords: Machine learning, active learning, enzyme promiscuity
Introduction
Substrate-level enzyme promiscuity(Humble and Berglund, 2011; Khersonsky and Tawfik, 2010; Sévin et al., 2016) has recently been recognized as a far more ubiquitous phenomenon than previously assumed, having important implication in several research areas. Substrate-level enzyme promiscuity is a phenomenon in which an enzyme can catalyze a reaction on more than one substrate, and the challenges it presents in experimental methods have made development of in silico methods for its prediction a subject of intense interest. Understanding this phenomenon is critical to explaining many biological processes. Enzyme promiscuity plays a key role in metabolite damage(Linster et al., 2013), a phenomenon where essential metabolites are converted to non-useful forms by reactions catalyzed by both homologous and heterologous promiscuous enzymes in wild-type and engineered organisms. Metabolite damage can drastically reduce fitness, to the extent that specific repair mechanisms have evolved to convert damaged metabolites back into a form that can be used by the cell(Linster et al., 2013; Van Schaftingen et al., 2013). Enzyme promiscuity is also critical in specific mechanisms of antibiotic resistance. Promiscuous enzymes can compensate for inhibited essential enzymes, allowing bacteria to circumvent the block. For example, methotrexate is an effective antibiotic against many microbes by inhibiting dihydrofolate reductase, an essential enzyme. However, in Lieshmania major, a second enzyme, PRT1, a broad spectrum pteridine reductase, is able to catalyze the DHFR reaction and is not inhibited by methotrexate(Gourley et al., 2001; Nare et al., 1997). Finally, enzyme promiscuity is important in metabolic evolution: It has been hypothesized that promiscuous enzymes improve fitness by serving as a starting point in biochemical evolution(DePristo, 2007; Khersonsky and Tawfik, 2010). This would explain instances of related proteins having a wide range of activities(Verdel-Aranda et al., 2015) and the presence of pathways that rescue what would otherwise be lethal knockout strains(Kim and Copley, 2012).
Enzyme promiscuity also has substantial positive and negative effects on industrial biotechnology. Substrate-level promiscuity can enable a tantalizing array of novel biosynthetic routes to drugs and biochemical. Promiscuous enzymes may also catalyze non-canonical reactions, allowing for the potential construction of metabolic routes to compounds not known to occur in nature. On the other hand, enzyme promiscuity can also be problematic to industrial biotechnology applications. Heterologous enzymes can promiscuously act on unanticipated native metabolites, diverting carbon from a desired end product(Mafu et al., 2016); in engineered pathways, concentrations of heterologous enzymes and metabolic intermediates are pushed to high concentrations such that the probability of substrate-level promiscuity is high, leading to unintended and deleterious effects on biochemical production (Biggs et al., 2016). To address this, there has a been a wave of computational approaches to design novel metabolic pathways and predict byproduct-producing and/or damage reactions (Campodonico et al., 2014; Carbonell et al., 2014; Cho et al., 2010; Lee et al., 2012). A rising challenge in applying these methods is low accuracy, largely due to a paucity of high-quality data on which to best train in silico prediction models.
Enzyme promiscuity is commonly investigated by assaying the activity of an enzyme on several different compounds selected ad hoc. However, selecting substrates that best expand the knowledge about an enzyme’s promiscuity is a non-trivial task, as there are a large number of possible substrates. Furthermore, several compounds may be prohibitively expensive or difficult to procure, making an exhaustive experimental study infeasible. To avoid the time and expense of carrying out activity assays, in silico methods can be substituted that either make use of existing promiscuity data or reduce experimental effort by collecting informative substrates. Promiscuity characterization is a task that can be addressed by cheminformatics and machine learning methods. While molecular modeling suites and docking approaches are capable of providing more nuanced predictions about the interactions between enzymes and potential substrates than 2D cheminformatics approaches, crystal structure information they require is often not available.
In order to predict enzyme promiscuity, previous studies(Campodonico et al., 2014; Cho et al., 2010; Pertusi et al., 2015) have deployed similarity-based approaches to predict an enzyme’s ability to catalyze a reaction on a given substrate. These methods rely on catalogues of known substrates as listed in databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG)(Kanehisa et al., 2014) or the Braunschweig Enzyme Database (BRENDA)(Schomburg et al., 2013). Similarity methods (Willett, 2006) and supervised machine learning methods are subsequently used to rank and/or classify likely substrates, and has been applied to numerous protein targets and off-targets in drug development efforts (i.e., CYP450 (Jacob and Vert, 2008; Terfloth et al., 2007; Wale and Karypis, 2009)). Support vector machines(Schölkopf and Smola, 2002) (SVMs) are an approach that has performed well in these studies and are trained on data belonging to each of two labeled classes of interest. In the context of predicting substrate-level enzyme promiscuity, the labeled classes are enzyme substrates and non-substrates. The resulting model can then be used to predict if an untested compound is a substrate or non-substrate (cf. Methods). With a previous study estimating that 39% of enzymes in KEGG exhibit substrate promiscuity (Carbonell and Faulon, 2010), the extension of this approach to metabolic enzymes can streamline the process of identifying promiscuous enzymes with a desired side activity as a prelude to structure-based engineering efforts.
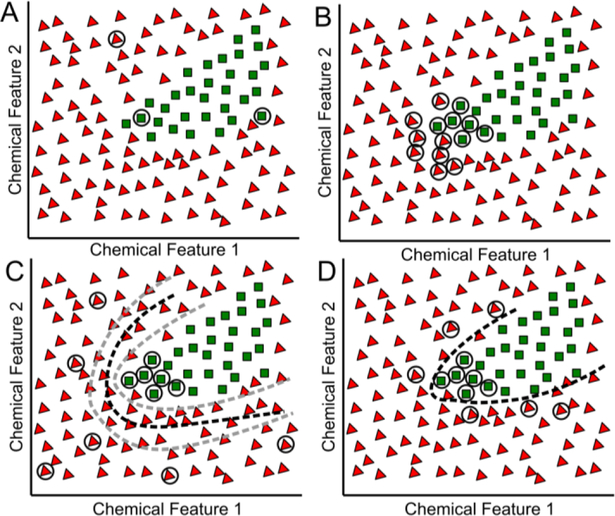
Existing databases used to train enzyme promiscuity models have limitations. For most enzymes of biosynthetic interest, the datasets of tested compounds are not very large. As an example, only ~5% of enzymes across all Escherichia coli strains in the BRENDA database have 20 or more reported substrates (e.g., Figure 1A, Table S1). Secondly, these datasets contain disproportionately small numbers of inactive compounds, without which the task of distinguishing between substrates and inactive compounds is highly uncertain (Figure 1C, 1D). Finally, existing datasets do not generally explore a diverse set of possible substrates may contain many compounds of relatively low diversity, and therefore low information content (Figure 1B). While there is no consensus on the number of training compounds required to adequately train an SVM classifier, it is clear that larger, more diverse compound datasets will be required to improve classification power(Matykiewicz and Pestian, 2012), and collecting this data efficiently is very important.
Figure 1. Challenges in characterizing enzyme promiscuity.
Existing datasets of active compounds (green squares) and inactive compounds (red triangles) describing substrate-level enzyme promiscuity often consist of (A) narrow distributions in chemical space or (B) a small number of compounds. (C) In the absence of negative data, randomly selected compounds used in its stead can be widely dispersed, leading to a high false positive rate when used to train SVMs due to high uncertainty in the position of the decision surface (dashed lines). (D) By comparison, confirmed inactive data near the decision surface allows for less uncertainty in calculating an optimal separating hyperplane.
In order to probe chemical space in an efficient manner while still avoiding the drawbacks of exhaustive experimentation, an active learning method can be employed to direct the task of data collection. Active learning(Settles, 2012; Warmuth et al., 2003) is a term applied to a number of approaches that use information about the set of unlabeled instances (i.e., untested compounds) in order to make strategic choices about which unlabeled instances to query next so that the result can efficiently train the classifier. In the case of SVMs for predicting substrate-level enzyme promiscuity, active learning serves to prioritize compounds in chemical space according to the additional predictive power knowing the result of that substrate may add to the SVM. Active learning could then offer a roadmap to experimentally efficient enzyme characterization. However, a question remains as to whether this approach can provide sensible compounds as suggestion despite the relatively small datasets available.
In this study, we examine the utility of SVM-based machine learning to predict enzyme substrate promiscuity across a range of enzymatic chemistries and examine the ability of active learning to prioritize new compounds for testing that efficiently expand the domain of applicability of the classifier. Specifically, we compile four enzymes’ datasets and develop SVMs for each of them using existing data, demonstrating their efficacy on relatively homogenous chemical sets. We then develop an active learning approach to strategically expand the available pool of compounds—both active and inactive—to use as training data in developing SVM classification models for metabolic enzymes. To our knowledge, we are the first to apply active learning to metabolic enzymes and demonstrate that the approach is effective. We use SVM classification models to validate the efficacy of the active learning method by cross validating existing data. We go on to use an active learning approach to rank untested compounds containing the putative molecular active site from the ZINC Is Not Commercial (ZINC) database(Irwin et al., 2012)—a diverse catalogue of biochemically relevant chemicals—as to their ability to add classifying power to the model for a case study enzyme. Finally, we demonstrate that highly-ranked compounds are enriched for chemical features that add chemical diversity to the dataset.
Results
Existing Datasets Have Inadequate Information Content to Evaluate Substrate-Level Promiscuity
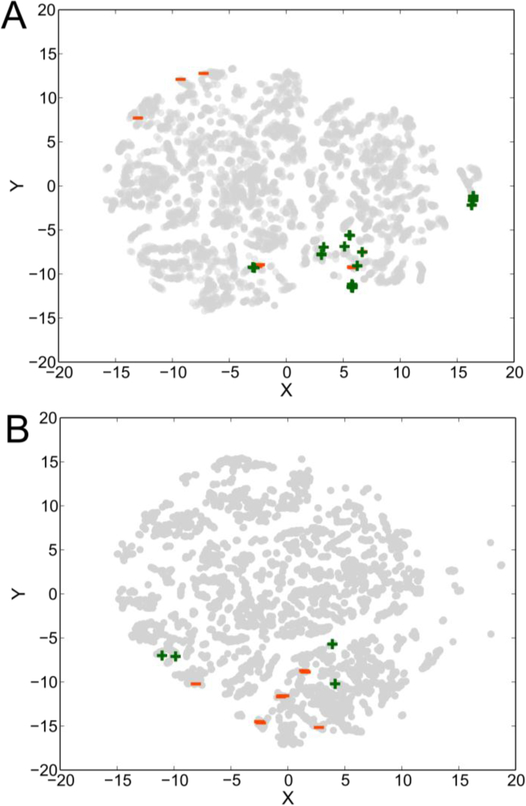
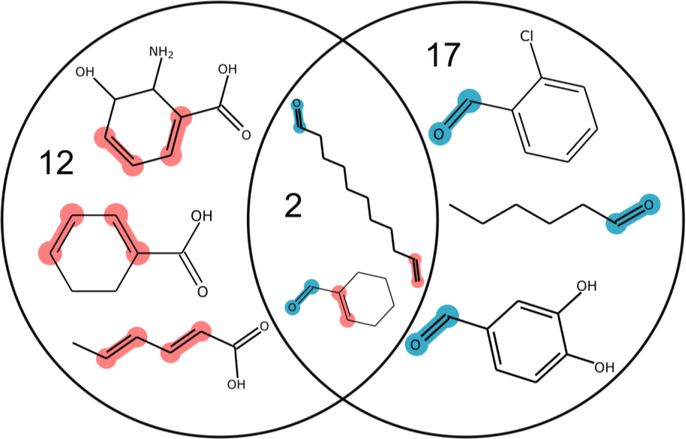
Datasets with diverse chemical features are essential to predictive SVMs, as datasets containing only highly similar chemicals do not lead to accurate predictions on chemicals that are very different from the training set. Diverse sets of chemicals, however, make more accurate predictions for a wider set of potential substrates. To visualize the diversity of existing substrate-level promiscuity data, we first generated t-distributed stochastic neighbor embeddings (tSNE, cf. Methods) for each of the four compound datasets for the four enzymes considered in this study: 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid synthase (MenD) from Escherichia coli(Kurutsch et al., 2009), carboxylic acid reductase (Car) from Nocardia iowensis(Akhtar et al., 2013; Moura et al., 2016; Venkitasubramanian et al., 2008, 2007a, 2007b), amino acid ester hydrolase (AAEH) from Xanthomonas citri(Kato et al., 1980), and 4-hydroxyacetophenone monooxygenase (HAPMO) from Pseudomonas putida(Rehdorf et al., 2009) (Figure 2, S2). This embedding reduces the many-dimensions in which chemicals vary to a two-dimensional projection while preserving the property that highly similar chemical structures will generally cluster closely together in the resulting visualization. In the case of the Car dataset, we noted that there is one larger cluster that dominates (Figure 3A), with a small number of outlier compounds. This result is representative of the scenario described in Figure 1B, and indicates that models generated with this dataset are unlikely to provide accurate results for more varied compounds. On the other hand, in the case of MenD (Figure 3B), we noted that there also markedly distinct clusters of compounds, though not well distributed. We also evaluated two more enzymes, HAPMO and AAEH. We observed that the HAPMO data set (Figure S3) was more evenly distributed, but in a relatively small chemical space, similar to Car. The AAEH embedding (Figure S4) showed a starker separation between putative substrates, with compounds located primarily in tight clusters within each substrate class (amino acid vs. antibiotic). Based on this analysis, we would anticipate that SVMs for MenD and AAEH would have a higher accuracy when making predictions on similar compounds to those in the dataset, as these models account for more chemical features, whereas the Car and HAPMO data would result in models with potentially higher global performance—though performance on cross-validation could be diminished due to a lack of representative chemicals from any given cluster.
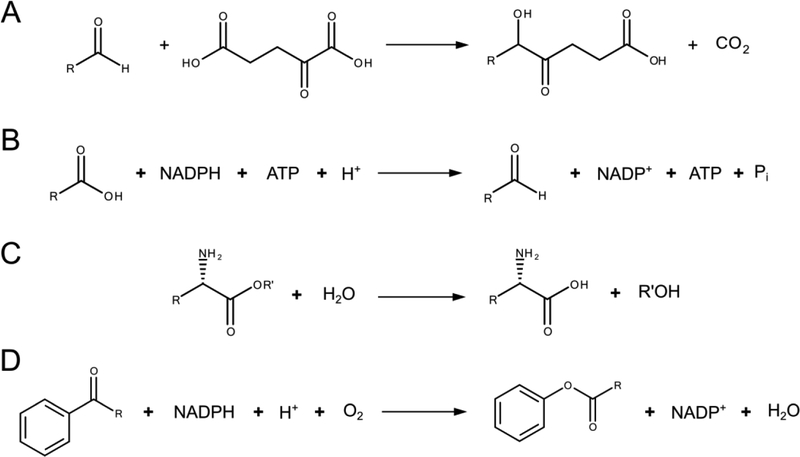
Figure 2. Representative reaction schemes for the enzymes analyzed in this study.
Schemes for (A) MenD and (B) Car are conserved for each reaction known to be catalyzed by these enzymes. The reactions catalyzed by (C) AAEH and (D) HAPMO allow for more structural diversity. In particular, AAEH can also cleave at a peptide bond with similar local structure to the one in this figure, and HAPMO may oxygenate at sites in a cyclic aliphatic system adjacent to a carbonyl.
Figure 3. Compounds selected by scientists are not necessarily diverse.
tSNEs for (A) Car and (B) MenD. Within each set, there are multiple distinct portions of chemical space represented, yet the existing datasets do not capture the diversity inherent in biologically relevant chemical space. Active compounds in each set are represented by green +, inactive compounds by red −, and untested compounds by grey circles.
Next, we determined predictive performance using the datasets in a cross-validation of SVM models for the enzymes in question. For each dataset, we sampled ~60% of the molecules for training an SVM, using the remaining compounds as a test set to measure the accuracy of the model. We repeated this procedure 1,000 times for each enzyme and calculated a final estimated accuracy by averaging the results for each dataset (). For the Car data set, we observed an of 75% ± 0.6%; for MenD, approximately 79% ± 0.8%; for HAPMO, approximately 76% ± 0.6%; and for AAEH, approximately 81% ± 0.6%. As expected, the more diverse sets performed somewhat worse on the held-out data.
For each of the models, we desired to see if the chemical features that had the greatest importance to the model were in concordance with biochemical knowledge of the datasets. For this, we generated SVMs using the full datasets for each enzyme and calculated the impact each chemical feature had in distinguishing between active substrates and inactive substrates for each enzyme. These results are summarized in Table 1 and demonstrate that our models identify that a small number of features contribute to the decision-making. For example, carbon-carbon double bonds and aldehydes are considered highly important in the case of the MenD classifier, as both of these features are highly correlated with membership in the active substrates class. The Car classifier identifies nitrogen-containing groups as most discriminatory for much the same reason—these features are generally found in the inactive compounds.
Table 1.
Scores for the most informative features in SVMs constructed from existing datasets, shown for comparison alongside the features that are most enriched for in compounds prioritized by active learning, with p-values and effect sizes based on Barnard’s exact test at a significance level of α = 0.05, adjusted with the Bonferroni correction.
| Car | ||||||
|---|---|---|---|---|---|---|
| Base SVM Feature Scores | Enriched Features | p-value | Effect Size | −95% CI | +95% CI | |
| amine | 0.13 | 1,3-tautomers | 1.15 × 10−10 | 0.120 | 0.073 | 0.166 |
| α-amino acid | 0.13 | R chirality | 4.37 × 10−6 | 0.089 | 0.049 | 0.130 |
| nitro | 0.04 | 2° amide | 5.36 × 10−6 | 0.070 | −0.008 | 0.148 |
| 1° aliphatic amine | 0.04 | 2° alcohol | 2.61 × 10−5 | 0.072 | 0.006 | 0.138 |
| 1,3 tautomers | 0.04 | amide | 2.62 × 10−5 | 0.069 | 0.003 | 0.135 |
| MenD | ||||||
| Base SVM Feature Scores | Enriched Features | p-value | Effect Size | −95% CI | +95% CI | |
| aldehyde | 0.14 | C=C bond | 4.87 × 10−65 | 0.509 | 0.448 | 0.570 |
| C=C bond. | 0.13 | Michael acceptor | 1.16 × 10−54 | 0.462 | 0.401 | 0.524 |
| Michael acceptor. | 0.11 | cis double bond | 2.26 × 10−25 | 0.291 | 0.218 | 0.364 |
| rotable bond | 0.09 | trans double bond | 2.26 × 10−25 | 0.291 | 0.218 | 0.364 |
| 1,5 tautomers | 0.07 | 1,3-tautomers | 7.91 × 10−18 | 0.212 | 0.153 | 0.271 |
| HAPMO | ||||||
| Base SVM Feature Scores | Enriched Features | p-value | Effect Size | −95% CI | +95% CI | |
| rotable bond | 0.25 | rotable bond | 1.17 × 10−6 | 0.173 | 0.086 | 0.260 |
| AAEH | ||||||
| Base SVM Feature Scores | Enriched Features | p-value | Effect Size | −95% CI | +95% CI | |
| amine | 0.21 | aromatic | 2.07 × 10−40 | 0.614 | 0.517 | 0.712 |
| 1° amine | 0.21 | ortho substituted | 2.52 × 10−13 | 0.318 | 0.215 | 0.421 |
| aromatic | 0.07 | meta substituted | 6.10 × 10−10 | 0.277 | 0.170 | 0.384 |
| conjugated dbl bond | 0.03 | hetereoaromatic | 2.51 × 10−9 | 0.272 | 0.170 | 0.376 |
| 1,3 tautomers | 0.02 | 2° mixed amine | 3.10 × 10−9 | 0.258 | 0.141 | 0.375 |
Active Learning Is an Effective Approach for Efficiently Sampling Diverse Chemical Spaces
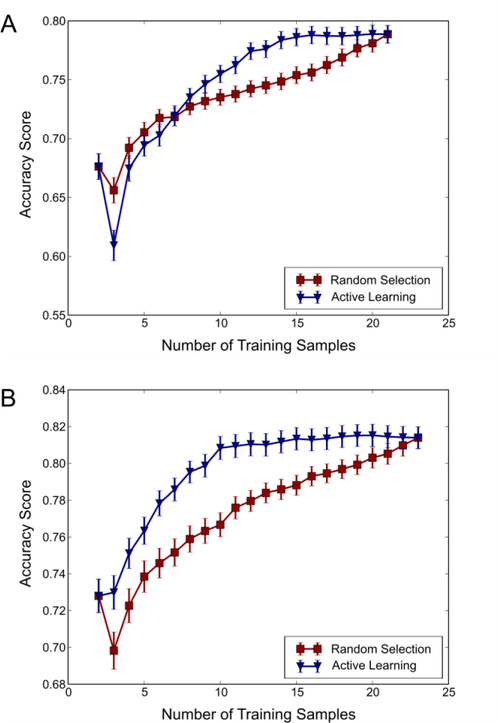
Smaller compound datasets with a limited number of chemical features can make predictions only about related compounds, and are the majority of enzyme promiscuity studies to date. Since probing chemical space in order to characterize enzymes can be costly and time consuming, we next evaluated the performance of active learning to increase predictive power with minimal experimental burden. To ascertain the benefit of sampling a larger cross-section of chemical space using an active learning technique, we first performed an internal assessment of the existing datasets for MenD and AAEH, as the datasets would allow active learning to sample moderately diverse compounds. In practice, active learning would select compounds from a large commercially available library, but for initial validation, we used existing data that could be easily cross validated. The learning curves were generated as described in the Methods, tracking the increase in classifier accuracy as additional compounds were added to the training set. This is analogous to investigating the accuracy gain as experiments are carried out on new compounds, and including that information in the training set. We expected SVMs whose training data were chosen using an active learning method to approach more quickly than models whose datasets were selected at random. The learning curves for both MenD and AAEH exhibited the hypothesized behavior: Despite the small datasets, a significantly faster gain in accuracy was observed for datasets assembled using an active learning method when compared to random selection in both cases (Figure 4). In both cases, we observed that the value of was reached with a significantly smaller number of compounds than were available for selection. In the case of MenD, this value was reached with approximately 30% fewer compounds, while in the case of AAEH, the maximum average accuracy was reached with approximately 50% fewer compounds than were available for selection. We underscore that this would be a substantial savings in experimental effort, if active learning was used to guide the enzyme promiscuity characterization. We observed no significant advantage with an active learning approach in the cases of Car and HAPMO, presumably because these datasets were highly homogenous, with little diversity for active learning to sample (Figure S5). From this, it can be concluded that while chemical diversity is prerequisite for good prediction, achieving diversity using a substrate sampling method such as active learning is important to minimize the experimental burden.
Figure 4. Active learning improves model accuracy with significantly fewer compounds.
Learning curves for SVM models of (A) MenD and (B) AAEH. In both cases, the maximum accuracy of the classifier is reached when selecting compounds using active learning. Error bars represent one standard deviation from the mean value of the accuracy score calculated across 1,000 iterations.
Active Learning Adds Relevant Features and Structural Diversity
In the previous active learning study, the active learning algorithm was only allowed to sample from the small number of compounds that data was available for. In a practical application, active learning would rank and select from a substantially larger set of commercially available compounds. To test if active learning, when allowed to select from such a large database, would select compounds that contained chemical moieties likely to inform the model being developed, we allowed the algorithm to select compounds from the very large and diverse ZINC database. For each of the four case study enzymes, we queried the ZINC database for all compounds containing the relevant reactive atoms for the enzyme under investigation, limiting the results to those compounds whose mass was within one standard deviation of the average mass of the known active compounds. Using this criterion, 12,491 compounds were ranked as to their potential to add information to the classifier for Car; 5,542 compounds for the MenD classifier; 2,170 compounds for the AAEH classifier; and 5,252 compounds for the HAPMO classifier (Supplementary Files). In order to assess if the active learning-produced rankings were biased for informative chemical features, we counted the occurrences of each chemical feature accounted for in the chemical fingerprints in the top 10% of the compounds prioritized by active learning and compared it to the occurrence in an equal number of randomly selected compounds. We then tested for the significance of the presence of a particular feature in a compound set using Barnard’s exact test at a significance level of α = 0.05/N, where N is the number of features in the fingerprint and is used to make the Bonferroni correction. Notably, compounds in the top 10% of rankings generated with active learning were enriched for features both present and not present in the training sets (Table 1, S2).
It is possible that active learning is merely giving high weight to dissimilar compounds, rather than compounds that maximize information content. As we describe in the previous section, diverse sets are necessary but not sufficient to maximize the information content in enzyme substrate classification. For example, a compound could contain unique features, but the remaining features may already allow the model to make a confident classification as to that compounds reactivity. To test this, we compared the enrichment of features using an active learning method with any potential enrichment based solely on compound dissimilarity. In order to calculate the enrichment for a dissimilarity-based approach, we first calculated the centroids of the training sets. We then calculated the Tanimoto score (i.e., chemical similarity score) between the centroid and each of the compounds in the unlabeled set and sorted them such that compounds with the lowest Tanimoto score ranked highest. Using the dissimilarity search rankings, only features not present in the training data (aside from the features in molecular active site) were enriched for (Table S3) in the top 10% of the list, which greatly increases the chances of selecting outlier data over relevant data. Chemical space is high-dimensional, as there are typically more than 300 features considered for a compound set, making it likely that chemically different outlier compounds would be chosen over relevant, discriminatory compounds found by active learning.
To experimentally validate the approach, two sets of four compounds (1–4 and 5–8) were selected from the active learning-prioritized lists for MenD with the intent of performing activity assays with these compounds, adding the resulting data to the existing SVM model, and examining the effect. Compounds were selected that could be obtained in amounts and purities that were sufficient for performing the assays; 1–4 were chosen from the top 10% of high ranking compounds, while 5–8 were chosen from the bottom 90%. The activity assays revealed that all eight compounds could serve as substrates for MenD (Figures S6–S13). Notably, compounds 1–4 caused large shifts in importance scores for features previously known to be highly discriminatory (Table S4), as compared to the shifts induced by the introduction of data corresponding to compounds 5–8. Therefore compounds 1–4 were more informative about chemical space (represented by the ZINC compounds) than were compounds 5–8.
Finally, we sought to test if the new, more informed SVM model improved prediction. To test this, we identified compounds that were predicted to be negatives by the original SVM model, but were predicted to be positive using the refined SVM (including the new training data from compounds 1–8). 53 compounds previously not predicted as substrates for MenD were newly predicted to be promiscuous substrates. Of these, a few commercially available compounds (9–11) were tested and all were found to be substrates for MenD (Figures S14–S17), demonstrating the ability of this approach to effectively uncover new potential reactions.
Discussion
Understanding and predicting enzyme promiscuity is becoming increasingly important in both understanding metabolism and in designing novel metabolic pathways for biosynthesis. In each case, the enzyme-substrate data is vastly smaller than the number of promiscuous reactions we would like to consider. The large number of possible metabolic intermediates is vastly larger that the compounds available for purchase(Irwin et al., 2012; Lucas et al., 2015). Because many metabolic intermediates are not readily purchasable, the expense and time required for custom synthesis of compounds to test with an enzyme is at best highly undesirable, or at worse it is simply not amenable to synthetic techniques. Beyond compound availability, the importance of having a diverse set of compounds as training data becomes greater as interest shifts to the production of chemical classes with higher potential for structural variety, such as drugs. Machine-learning models allow us to take existing data and make predictions on untested compounds, substantially saving time and effort. These models are directly applicable for selecting candidates for site-directed protein engineering or biocatalysts for designing pathways.
While rich datasets exist for some enzymes, there are many more enzymes that have potential utility in metabolic engineering that do not have such datasets available. In this work, we present a methodology for rapidly characterizing an enzyme’s specificity (by means of a SVM model) by strategically selecting compounds that maximizes the information obtained from the experiments. Active learning is an attractive approach for populating diverse compound sets for a specific enzyme. Selecting compounds randomly or by a dissimilarity metric to test may select (a) compounds that are redundant and do not serve to improve the model or (b) compounds that are outliers and thus of little relevance to chemical space. Active learning takes both the existing model and structure of chemical space into account when selecting compounds, meaning that the model will gain information to improve its own accuracy as well as represent the multitude of unlabeled compounds to be classified by the model. As novel features are introduced in this method, the model can adapt to the new information and serve to direct inquiry in subsequent iterations. This is also dependent upon the structure of the initial dataset, which we have observed results in distinct behaviors in the test cases presented in this work.
The compounds selected by the active learning algorithm for MenD emphasize both obvious and less intuitive chemical features. In the analysis of existing data, activity was strongly associated with aldehydes, which is trivial, as it is the reactive functional group for MenD (Figure 5). Conversely, compounds with double bonds were mostly in the inactive pool. The unanswered question in the original data is “Do double bonds make a compound inactive or is it only the lack of an aldehyde?” To answer this, active-learning gave high ranks to aliphatic aldehydes that simultaneously contained double bonds and related features. Conversely, benzaldehydes derivatives were not selected by active learning, because the benzaldehyde derivatives in the training data were reactive, so additional testing in this feature class is unimportant. The only aromatics that were highly ranked were aromatic aldehydes that have groups not represented in the training set.
Figure 5. Active learning selects compounds that delineate how features impact activity.
The MenD training set has large numbers of compounds that contain either an aldehyde group (pink) or a carbo-carbon double bond (blue), and a comparatively small number of compounds that contain both. ZINC aldehydes with carbon-carbon double bonds cannot be easily classified because there is insufficient training data to resolve the high correlation of aldehydes with active compounds and carbon-carbon double bonds with inactive compounds. Numbers indicate the number of compounds in each group (aldehyde, C-C double bond, or both).
The AAEH classifier is based on training data that is split into distinct categories: amino acid esters and β-lactam antibiotics, consisting mostly of cephalosporins. As most amino acid esters in the training set are active, it is desirable for the algorithm to select compounds that may serve to resolve which features are important factors in determining cephalosporin activity. As such, the top scoring compounds in active learning are enriched for features associated with aromatic rings, which are common side groups in cephalosporins. The top scoring compounds are also enriched for secondary mixed amines, which is a feature also common to β-lactam antibiotics.
In the case of Car, active learning results appear promising when allowed to select commercially purchasable compounds from ZINC, but had little benefit in the validation study where the algorithm could only select among a relatively homogenous set of compounds. The latter result is consistent with expectations. Upon analyzing the active learning results from ZINC for Car, we observed a significant enrichment in the top ranking results for tautomerizable compounds, with weaker enrichment for amides and alcohols. None of these features correspond directly to those that rank as highly discriminatory in the base classifier, consistent with the observation that these discriminatory features are distributed only in the negative training data. Finally, as the training data are rich in aromatic compounds, the significant lack of aromatic compounds in the top ranked active learning results demonstrates that the algorithm is assigning higher significance to understanding features associated with aliphatic compounds.
Finally, the active learning results generated for HAPMO exhibited no significant enrichment for any feature except rotable bonds. As rotable bonds are common in both the active and inactive compounds, the SVM cannot discern when rotable bonds are deterministic of activity. By prioritizing compounds that contain rotable bonds, the algorithm might provide other chemical features to distinguish between putative active and inactive compounds. The results also show a significant de-enrichment for compounds containing chemical features that are well-represented in the training set. The reduction in compounds with conjugated bonds and related features (not all conjugated ring systems need be aromatic or anti-aromatic) is indicative of a focus on introducing more varied structures.
The active learning algorithm was experimentally tested by purchasing four highly ranked compounds and four compounds from the lower echelons of the rankings and testing them for activity with MenD. The very highest ranked compounds from ZINC were only “purchasable” through custom synthesis, therefore, four compounds ranked lower in the list, but still in the top 10% were selected (1–4). In a practical application, using a compound database that contains only off-the-shelf compounds—and perhaps only those below a cost threshold—would be ideal. Compounds from the lower portion of the ranking (5–8) were also tested, with the expectation that they would contribute minimally to the types of predictions made by the model. This is reflected in the updated feature importance scores (Table S4), which demonstrate little change between the base classifier and the classifier with 5–8 added. Adding data for 1–4 to the training set, however, causes marked shifts in feature importance, chiefly in reducing the importance of the double bond feature. This is expected, as carbon-carbon double bonds were strongly correlated with inactive compounds in the initial training set. The introduction of new information about carbon-carbon double bond-containing compounds that serve as active substrates for MenD implies a more even distribution of that feature among the two classes, which reduces its discriminatory power and its importance score. Ultimately, this enhanced discrimination facilitated the identification of three novel MenD substrates that were previously not predicted to be catalyzed by the enzyme.
Besides reducing the number of compounds, the per-compound cost is another aspect that could be considered using active learning. Even for off-the-shelf compounds, cost can vary by orders-of-magnitude. From an active learning standpoint, information content of a molecule is not necessarily associated with cost, and selecting inexpensive compounds may train the model as well as expensive compounds. As an example of the potential savings, we looked to the learning curves for the existing Car data. In this case, across 1,000 iterations, the accuracy growth quickly flattened (Figure S5), suggesting the presence of several redundant compounds in the training set. We gathered prices for the smallest quantity of each compound in the training set greater than 1g, with an average cost per compound of approximately $47, with the most expensive being greater than $100 for the smallest quantity. As the set has a large amount of redundancy, the elimination of the five most expensive compounds from the dataset would result in minimal accuracy loss, while eliminating nearly 30% of the total expenditures for compounds—not to mention fixed costs associated with labor and analytical equipment. In the case of MenD, the asymptote was approximately 6 compounds long (Figure 3A), representing approximately 20% of the dataset. While pricing information for all of the compounds in this set was not available, compounds analogs were very expensive (>$500/g). We would anticipate the savings from eliminating these compounds from the set would be equal to or greater than those observed in the case of Car. By selecting compounds strategically using active learning, it may be possible to elide much of this redundancy in order to efficiently characterize enzymes of interest.
To exploit machine learning approaches to enzyme promiscuity requires both data for active substrates and inactive compounds, as the inactive compounds can be as informative as active substrates. To date, public databases do not efficiently compile reports of molecules that have been tested against enzymes of interest but have shown no activity (i.e., negative training data). Often, presumptive negative training data are assembled by random selection. While this approach can yield apparently good results on cross-validation(Heikamp and Bajorath, 2013), the separation in feature space between a randomly selected negative training set and a positive training set that may consist only of a tight distribution of compounds is likely to be large, leading to high false negative or false positive rates when used to classify further compounds (Figure 1C, 1D). A possible compromise approach to generating more accurate SVMs may therefore lie in endeavoring to expand the available training data in such a way that it becomes more akin to the background distribution of chemicals while remaining relevant to the enzyme that it is being constructed for(Smusz et al., 2013). Moreover, recent advances in high-throughput metabolomics techniques (Sévin et al., 2016) can be complemented by an active learning approach by enabling judicious selection of chemical matter to be screened, enabling rapid accumulation of data for model building on a wide range of enzymes. The method could be easily modified to accumulate active compounds rather than high information compounds by introducing weak reinforcement (Maciejewski et al., 2015).
The focused nature of many studies on enzyme activity has tended to produce enzyme-substrate activity data that is very much localized to a small sector of the available chemical space. While such specific data may serve individual studies quite well, the more localized nature of the datasets is incompatible with the goal of producing globally predictive activity models, the type of models that will be required to access the effects of promiscuity in metabolic networks. The active learning strategy represents an opportunity to systematically gather information about an enzyme’s substrate specificity that can subsequently be used to generate globally relevant models. Deploying such a method on a large scale is a task that can be aided by the advent of high-throughput techniques for enzyme characterization and web services that allow for the pooling of data. In terms of metabolic engineering, this approach can allow for efficient elucidation of the scope of the substrate promiscuity of enzymes of interest. The global datasets in turn can enable (1) the rapid, facile selection of enzymes for deployment in biosynthetic pathways; (2) the prioritization of candidate enzymes for protein engineering based on their latent promiscuity; and (3) the identification of enzymes whose promiscuous activity may explain observed microbial phenotypes.
Methods
Data Sources
Substrate promiscuity data were obtained from a search of the BRENDA online enzyme database(Scheer et al., 2011), and the corresponding published datasets. Positive training data consists of compounds listed as substrates of an enzyme from one organism from a BRENDA entry for one EC number, with additional training data added from literature and experiments as needed. Negative training data not listed in BRENDA consisting of inactive compounds were obtained from a manual search of the primary literature. Where possible, we selected negative training compounds containing the functional group required by the enzymatic activity under investigation, but include other tested inactive compounds when available, as these confirmed compounds still offer a greater degree of certainty than randomly selected compounds. The enzymes we investigated were 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid synthase (EC 2.2.1.9, MenD) from Escherichia coli(Kurutsch et al., 2009), carboxylic acid reductase (EC 1.2.1.30, previously 1.2.99.6, Car) from Nocardia iowensis(Akhtar et al., 2013; Moura et al., 2016; Venkitasubramanian et al., 2008, 2007a, 2007b), amino acid ester hydrolase (EC 3.1.1.43, AAEH) from Xanthomonas citri(Kato et al., 1980), and 4-hydroxyacetophenone monooxygenase (EC 1.13.14.84, HAPMO) from Pseudomonas putida(Rehdorf et al., 2009). Compounds ranked for active learning were taken from the ZINC(Irwin et al., 2012) database, version 12.
Plasmid Construction
The MenD gene was amplified from genomic E. coli DNA and cloned into a pET21a plasmid with the stop codon replaced by a four residue AALE peptide linker from the original pET21 construct, followed by a C-terminal 6×-His tag. All constructs were confirmed via DNA sequencing. DH5α E. coli was used for plasmid assembly, and the sequence-confirmed construct was transformed into BL21 (DE3) E. coli for protein expression.
Protein Expression and Purification
Two liters of MenD-expressing BL21 E. coli were grown in TB media with ampicillin at 37°C to an OD ≈ 0.6 in shake flasks. Expression was induced with 0.4 mM IPTG and grown overnight at 20°C for protein expression. Culture was spun down and re-suspended in 100 mL lysis buffer/g cells (50 mM phosphate, 20 mM imidazole, 150 mM NaCl, 1 mM Thiamine Diphosphate (ThDP), 1 mM MgCl2). The solution was lysed with an EmulsiFlex-C5 homogenizer (Avestin, Ottawa, Canada) at 15000 psi (pulsed) three times. Lysate was spun down at 18,000 rpm in a Beckman-Coulter (Brea, CA) JA-25.50 rotor (39,000 × g) for 40 minutes and the soluble fractions decanted for purification.
Protein purification of the soluble fraction was done with an AKTAExpress (GE, Pittsburgh, PA) protein purification chromatography system. The separation was done with a Hi-Trap FF Ni2+ column (GE, Pittsburgh, PA). The protein was eluted from the column with elution buffer (20 mM phosphate, 500 mM NaCl, 250 mM imidazole) and separated into nine fractions for SDS-PAGE analysis, which all showed high protein purity. Elution fractions were combined for buffer exchange and enzymatic assays. The purified protein in elution buffer was buffer exchanged three times into a storage solution (50 mM phosphate, 0.1 mM ThDP, 2 mM MgCl2) with Amicon 10 kDa cutoff centrifugal filters (Millipore, Billerica, MA). In the final solution, protein concentration was measured using a BioSpecNano (Shimadzu, Columbia, MD) with an extinction coefficient of 1.725 (mg/mL)−1cm−1 to be 2.4 mg/mL. The solution was split in half: one half was stored at 4°C for immediate use for assays with compounds 1–4, while the other was mixed with 10% glycerol, flash-frozen in liquid nitrogen, and stored at −80°C for subsequent assays with compounds 5–8 (Figure A in S1 Supporting Information). Positive controls confirmed enzymatic stability.
Enzymatic Assays for Compounds 1 - 8
Reaction solutions were prepared with 50 mM phosphate, 2 mM MgCl2, 0.1 mM ThDP, 50 mM 2-oxoglutarate, pH adjusted to 8 with NaOH, and stored at −20°C until use. For assays, reaction solutions were thawed to room temperature and 20 mM aldehyde substrates were added. Compound 1 was purchased from Hit2Lead; compound 4 was purchased from Acros Organics; compounds 3 and 6 were purchased from Alfa-Aesar; and the remaining four were purchased from Sigma-Aldrich (Figure A in S1 Supporting Information). MenD was added to a concentration of 0.133 mg/mL to initiate the reactions. Reactions were run in triplicate and BSA negative controls were also prepared for all reaction conditions. Reaction volumes of 2.0 mL were run at 30°C on a shaker platform in microcentrifuge tubes.
For reactions with poorly soluble substrates in the first set of compounds 1–4, insoluble particles were prior to removing the sample volume for GC/MS preparation to accurately evaluate the total amount of substrate left in each reaction vessel. During reactions with poorly soluble substrates in the second set of compounds 5–8, two sets of experimental conditions were tested for each substrate – one aliquotted with and one aliquotted without insoluble particulates after preparing the reaction solutions. The solution with no insoluble particles was used to monitor substrate consumption accurately, while the solution with insoluble particles was used as a semi-batch type system to allow for stronger putative product peaks (as more substrate was consumed, more un-dissolved substrate would go into solution to replace it). After the reaction, the solutions with insoluble particles were sampled from the supernatant only. The “dirty” solutions were sampled from the supernatant only. Aliquots of 500 μL were separated from the independent reaction vials and prepared for GC/MS at 0, 2, and 25 hours.
GC/MS Sample Preparation and Analysis
For each time point, small volumes (either 200 or 500 μL) of the total reaction volume were separated from the reaction vessels and 0.1% formic acid was added to the preparation samples to protonate putative products. These solutions were extracted into an equal volume of ethyl acetate. Aliquots of 400 μL of the ethyl acetate phase were separated for GC analysis. An internal standard solution of toluene was prepared and mixed with all sample solutions by addition of no more than 10% of the total volume. Standard curves for the substrates were also prepared in ethyl acetate ranging from 0.1 mM to 20 mM, with equivalent amounts of internal standard.
Gas chromatography was run on an Agilent 7890 GC with an HP-5MS-UI column (Agilent, Santa Clara, CA) with Tinlet = 250°C and a 15°C/min temperature ramp from 80°C to 325°C, except in the case of compound 4, where 45°C was used as a starting point. Mass Spectrometry was conducted in an Agilent 7000 QQQ in scan mode using Electron Ionization (EI). All samples were run at a 1:100 split ratio, and the 25 hour MenD time points were also run a second time at a 1:5 split to allow for detection of trace products.
Reactions were considered successful if the concentration of the aldehyde substrate in the MenD triplicate samples was significantly lower than the BSA negative controls at 24 hours. For substrates that showed significant depletion at 25 hours, product peaks were identified. In many cases, at least two potential product peaks appeared. These multiple peaks likely correspond to lactone and/or 2-oxo molecular products that arise during the sample preparation process and are the result of a successful 1,2 addition reaction(Kurutsch et al., 2009).
Enzymatic Assays for Compounds 9 - 11
Follow up enzymatic reactions were performed similarly for 9–11, but with negative controls consisting of all reaction components except the MenD enzyme, with extra buffer added to make up the volume, and with total reaction volumes of 1.5 mL. After running ~24 hours at 30°C, 300 rpm, reactions were split in two (750 uL) and extracted with 500 uL of ethyl acetate. Mixtures were vortexed and spun down in a tabletop microcentrifuge at max speed (17,000×g) for 10 min. Supernatant was collected and evaporated, and samples were re-suspended in 100 uL ethyl acetate and run on GC/MS. Compound 9 was obtained from Sigma-Aldrich; compound 10 was purchased from Combi-Blocks; compound 11 was purchased from Enamine.
Preprocessing Chemical Structure Data and Clustering
Compound names were converted to SMILES representations using the MolConvert Calculator Plugin(ChemAxon, 2013a), and Standardizer(ChemAxon, 2013b) was used for structure canonicalization and transformation. InChI representations of molecules were generated and read using the RDKit open-source cheminformatics platform (http://www.rdkit.org). The string representations of the molecules in the training set were converted to 2D chemical fingerprints with integer counts for use in developing machine learning models. Molecules were fingerprinted using the SMARTS(Daylight Inc., 2011) queries available in the OpenBabel(O’Boyle et al., 2011, 2008) FP4 fingerprint implementation, with additional queries for Cahn-Ingold-Prelog chirality assignments being carried out using RDKit. As we did not consider cofactors in the reactions, cofactors were removed from the dataset using a hand-curated set of cofactor pair rules. The centroid of the training sets is defined as the average of all the fingerprints in the set. The tSNEs generated for this study were generated using the scikit-learn and matplotlib Python libraries. The t-SNE algorithm is a stochastic clustering method(van der Maaten and Hinton, 2008) that attempts to preserve distance between data points in a higher-dimensional space while embedding it in a lower dimensional space. As a rule, proximal compounds are more similar than distant compounds, though distances between distant clusters of similar compounds may not be comparable due to the stochastic nature of the algorithm.
Predicting Substrates Using Support Vector Machines
The support vector machine (SVM) is a supervised machine learning method that can be used to classify compounds as being members of a class yi ∈ {+1, −1}, where +1 and −1 are labels denoting whether a compound is a substrate of an enzyme of interest or inactive with an enzyme of interest, respectively. To accomplish this, the SVM was first trained with data in the form of chemical fingerprints assembled in a matrix X, consisting of i fingerprints expressed as vectors of j features. The fingerprints in X can be thought of as vectors representing each compound that can be projected into a high-dimensional space. The training data was subsequently used to compute a decision function representing the distance from a hypothetical boundary dividing the two groups of compounds whose sign is used to predict the labels of molecules not in the training set. This function f calculated by quadratic program takes the following form, where x is a vector representing a query molecule; b is a threshold value determined from solving the quadratic program; xi and yi are the training compounds and their labels, respectively; αi are the Lagrange multipliers obtained from solving the quadratic program; and k(·,·) is a kernel function which computes a distance between two vectors(Schölkopf and Smola, 2002).
| (1) |
| (2) |
The choice of kernel function k allows the input data to be implicitly mapped into a high-dimensional feature space; this is desirable since there is likely to be non-linearity in the data. For this study, we select a radial basis function kernel:
| (3) |
Scikit-learn(Pedregosa et al., 2011), a machine learning package for Python, was used to generate the SVM models used in this study.
Identifying Discriminatory Chemical Moieties Using Feature Importance
The feature weights in SVM models making use of a non-linear kernel function such as the one in this study cannot be directly computed, but feature importance can be approximated by using recursive feature elimination(Mu et al., 2011) to calculate a ranking coefficient DJi, where α is the vector of Lagrange multipliers calculated from the full dataset, G is the Gram matrix calculated from the full dataset, and G−i is the Gram matrix calculated from the dataset with all counts for the ith feature in the fingerprint set to 0. Equation (4) can be used to calculate the effects of combinations of features by selecting multiple features i for which to set the counts equal to zero. The ranking coefficient can then be normalized by dividing it by the minuend in equation (4); this normalization then represents the percent change in the objective function of the SVM upon removal of a feature or set of features.
| (4) |
Active Learning
The ranking of compounds according to their ability to add predictive power to a base SVM model was achieved using an information density approach(Settles, 2012), which is a variety of active learning. Given a set of unlabeled compounds of interest , the information density score ζ of any given compound x in the set is calculated by equation (5), where U is the cardinality of the set :
| (5) |
| (6) |
| (7) |
Equation (5) requires a choice of uncertainty metric x, a similarity metric τ, and a tunable parameter β. In this study, ϕ was chosen to be the logarithm of the reciprocal absolute value of the decision function as in equation (6), τ was chosen to be the Tanimoto coefficient adapted for count fingerprints(Alvarsson et al., 2014) as in equation (7), and β was set equal to 1. Once compounds are ranked, the next instance is selected in a greedy fashion, queried (i.e., tested), and the result added to the dataset. The model is then retrained on the new dataset and the process is repeated until a stopping condition such as a target accuracy or dataset size is reached. In this study, we ran one iteration per enzyme, and performed activity assays on eight compounds in the active learning rankings for MenD so that a second iteration could be run. To reduce the computational burden, the ZINC database was filtered for compounds within 1 standard deviation of the average mass of the compounds in the positive training set for each SVM model in this study. We subsequently identified common molecular active sites in the positive training compounds and synthesized SMARTS queries to further filter the ZINC database to include in only those compounds containing a substructure capable of being reacted on by the enzyme of interest.
Learning curves are plots that demonstrate how the accuracy of an SVM grows as a function of the number of training examples used in developing it. These were generated for enzyme substrate datasets in this study to demonstrate the behavior of the accuracy of the SVM models as a function of the number of data points used to train them. The procedure for each enzyme was to divide the dataset into two parts: a training set, comprising 60% of the data selected at random, and a test set consisting of the remaining data. For each enzyme in this study, two SVM models, Λ and Ω, were used to generate the learning curves, where Λ is trained with data selected in an order prescribed by active learning Ω and is trained by selecting compounds at random. One data point from each label was randomly chosen, and these compounds (represented as fingerprints) were used to initially train both and . The training set was used to calculate an accuracy score a for the model, where t+ and t− are the number of compounds whose labels yi were correctly identified as +1 or −1, respectively and itest is the number of compounds in the test set:
| (8) |
At this point, the remaining compounds in the training set were added to the models one at a time. Compounds to be added to Λ were first scored according to the information density criterion in equation (5), and the highest scoring compound was selected. The compounds added to Ω were selected in random order. After each compound was added to the dataset, each model was retrained and a was calculated; this procedure was continued until the all compounds in the training set had been selected. This process was repeated for 1000 iterations, and an average value of a at each size of the training set was calculated. Curves for both models end on the same final value , since they are both trained on precisely the same dataset at the end of each iteration.
Supplementary Material
An algorithm for prioritizing experiments for enzyme promiscuity is proposed
The algorithm (SimAL) utilizes cheminformatics and support vector machine learning
SimAL can predict an enzyme’s promiscuity with 33% less experimental observations
SimAL judiciously resolve apparent conflicts in existing data in compound selection
Improvement in SimAL predictive power is experimentally validated
Acknowledgements
The authors would like to thank the Northwestern Recombinant Protein Production Core (rPPC) for protein expression and purification and NUSeq Core for sequencing services. Funding was provided by the NIH Biotechnology Training Program grant [T32-GM008449–23, D.A.P., B.W.B.]. the Bill and Melinda Gates Foundation [OPP1044008, M.E.M., K.E.J.T.], and the NSF [CBET50835800, M.E.M.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Akhtar MK, Turner NJ, Jones PR, 2013. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. PNAS 110, 87–92. doi: 10.1073/pnas.1216516110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarsson J, Eklund M, Engkvist O, Spjuth O, Carlsson L, Wikberg JES, Noeske T, 2014. Ligand-based target prediction with signature fingerprints. J. Chem. Inf. Model 54, 2647–2653. doi: 10.1021/ci500361u [DOI] [PubMed] [Google Scholar]
- Biggs BW, Rouck JE, Kambalyal A, Arnold W, Lim CG, De Mey M, Oneil-Johnson M, Starks CM, Das A, Ajikumar PK, 2016. Orthogonal Assays Clarify the Oxidative Biochemistry of Taxol P450 CYP725A4. ACS Chem. Biol 11, 1445–1451. doi: 10.1021/acschembio.5b00968 [DOI] [PubMed] [Google Scholar]
- Campodonico MA, Andrews BA, Asenjo JA, Palsson BO, Feist AM, 2014. Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm, GEM-Path. Metab. Eng 25, 140–158. doi: 10.1016/j.ymben.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Carbonell P, Faulon J-L, 2010. Molecular signatures-based prediction of enzyme promiscuity. Bioinformatics 26, 2012–9. doi: 10.1093/bioinformatics/btq317 [DOI] [PubMed] [Google Scholar]
- Carbonell P, Parutto P, Herisson J, Pandit SB, Faulon J-L, 2014. XTMS: pathway design in an eXTended metabolic space. Nucleic Acids Res. 42, W389–94. doi: 10.1093/nar/gku362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChemAxon, 2013a. Molecule File Converter.
- ChemAxon, 2013b. Standardizer.
- Cho A, Yun H, Park JH, Lee SY, Park S, 2010. Prediction of novel synthetic pathways for the production of desired chemicals. BMC Syst. Biol 4, 35. doi: 10.1186/1752-0509-4-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daylight Inc., 2011. SMARTS: A Language for Describing Molecular Patterns, in: Daylight Theory Manual. Daylight Chemical Information Systems, Inc., Laguna Niguel, CA, pp. 19–25. [Google Scholar]
- DePristo MA, 2007. The subtle benefits of being promiscuous: adaptive evolution potentiated by enzyme promiscuity. HFSP J. 1, 94–8. doi: 10.2976/1.2754665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourley DG, Schüttelkopf a W., Leonard G. a, Luba J, Hardy LW, Beverley SM, Hunter WN, 2001. Pteridine reductase mechanism correlates pterin metabolism with drug resistance in trypanosomatid parasites. Nat. Struct. Biol 8, 521–525. doi: 10.1038/88584 [DOI] [PubMed] [Google Scholar]
- Heikamp K, Bajorath J, 2013. Comparison of confirmed inactive and randomly selected compounds as negative training examples in support vector machine-based virtual screening. J. Chem. Inf. Model 53, 1595–1601. doi: 10.1021/ci4002712 [DOI] [PubMed] [Google Scholar]
- Humble MS, Berglund P, 2011. Biocatalytic Promiscuity. European J. Org. Chem 2011, 3391–3401. doi: 10.1002/ejoc.201001664 [DOI] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG, 2012. ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model 52, 1757–68. doi: 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob L, Vert JP, 2008. Protein-ligand interaction prediction: An improved chemogenomics approach. Bioinformatics 24, 2149–2156. doi: 10.1093/bioinformatics/btn409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M, 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–205. doi: 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Kawahara K, Takahashi T, Kakinuma A, 1980. Substrate Specificity of α-Amino Aicd Ester Hydrolase from Xanthomonas citri. Agric. Biol. Chem 44, 1075–1081. [Google Scholar]
- Khersonsky O, Tawfik DS, 2010. Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annu. Rev. Biochem 79, 471–505. doi: 10.1146/annurev-biochem-030409-143718 [DOI] [PubMed] [Google Scholar]
- Kim J, Copley SD, 2012. Inhibitory cross-talk upon introduction of a new metabolic pathway into an existing metabolic network. PNAS 109. doi:10.1073/pnas.1208509109/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1208509109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurutsch A, Richter M, Brecht V, Sprenger G. a., Müller M, 2009. MenD as a versatile catalyst for asymmetric synthesis. J. Mol. Catal. B Enzym 61, 56–66. doi: 10.1016/j.molcatb.2009.03.011 [DOI] [Google Scholar]
- Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY, 2012. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol 8, 536–46. doi: 10.1038/nchembio.970 [DOI] [PubMed] [Google Scholar]
- Linster CL, Van Schaftingen E, Hanson AD, 2013. Metabolite damage and its repair or pre-emption. Nat. Chem. Biol 9, 72–80. doi: 10.1038/nchembio.1141 [DOI] [PubMed] [Google Scholar]
- Lucas X, Grüning BA, Bleher S, Günther S, 2015. The purchasable chemical space: A detailed picture. J. Chem. Inf. Model 55, 915–924. doi: 10.1021/acs.jcim.5b00116 [DOI] [PubMed] [Google Scholar]
- Maciejewski M, Wassermann AM, Glick M, Lounkine E, 2015. An Experimental Design Strategy: Weak Reinforcement Leads to Increased Hit Rates and Enhanced Chemical Diversity. J. Chem. Inf. Model 150427115809005. doi: 10.1021/acs.jcim.5b00054 [DOI] [PubMed] [Google Scholar]
- Mafu S, Jia M, Zi J, Morrone D, Wu Y, Xu M, Hillwig ML, Peters RJ, 2016. Probing the promiscuity of ent-kaurene oxidases via combinatorial biosynthesis. Proc. Natl. Acad. Sci 113, 5–10. doi: 10.1073/pnas.1512096113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matykiewicz P, Pestian J, 2012. Effect of small sample size on text categorization with support vector machines, in: Proceedings of the 212 Workshop on Biomedical Natural Language Processing pp. 193–201. [Google Scholar]
- Moura M, Pertusi D, Lenzini S, Bhan N, Broadbelt LJ, Tyo KEJ, 2016. Characterizing and predicting carboxylic acid reductase activity for diversifying bioaldehyde production. Biotechnol. Bioeng 113, 944–952. doi: 10.1002/bit.25860 [DOI] [PubMed] [Google Scholar]
- Mu F, Unkefer CJ, Unkefer PJ, Hlavacek WS, 2011. Prediction of metabolic reactions based on atomic and molecular properties of small-molecule compounds. Bioinformatics 27, 1537–45. doi: 10.1093/bioinformatics/btr177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nare B, Hardy LW, Beverley SM, 1997. The Roles of Pteridine Reductase 1 and Dihydrofolate Reductase-Thymidylate Synthase in Pteridine Metabolism in the Protozoan Parasite Leishmania major. J. Biol. Chem 272, 13883–13891. doi: 10.1074/jbc.272.21.13883 [DOI] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James C a, Morley C, Vandermeersch T, Hutchison GR, 2011. Open Babel: An open chemical toolbox. J. Cheminform 3, 33. doi: 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle NM, Morley C, Hutchison GR, 2008. Pybel: a Python wrapper for the OpenBabel cheminformatics toolkit. Chem. Cent. J 2, 5. doi: 10.1186/1752-153X-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É, 2011. Scikit-learn: Machine learning in python. J. Mach. Learn. Res 12, 2825–2830. [Google Scholar]
- Pertusi DA, Stine AE, Broadbelt LJ, Tyo KEJ, 2015. Efficient searching and annotation of metabolic networks using chemical similarity. Bioinformatics 31, 1016–1024. doi: 10.1093/bioinformatics/btu760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehdorf J, Zimmer CL, Bornscheuer UT, 2009. Cloning, expression, characterization, and biocatalytic investigation of the 4-hydroxyacetophenone monooxygenase from Pseudomonas putida JD1. Appl. Environ. Microbiol 75, 3106–3114. doi: 10.1128/AEM.02707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D, 2011. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 39, D670–6. doi: 10.1093/nar/gkq1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölkopf B, Smola AJ, 2002. Learning with Kernels. The MIT Press, Cambridge, Massachusetts. [Google Scholar]
- Schomburg I, Chang A, Placzek S, Söhngen C, Rother M, Lang M, Munaretto C, Ulas S, Stelzer M, Grote A, Scheer M, Schomburg D, 2013. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 41, D764–72. doi: 10.1093/nar/gks1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles B, 2012. Active Learning, Synthesis Lectures on Artificial Intelligence and Machine Learning. Morgan & Claypool. doi: 10.2200/S00429ED1V01Y201207AIM018 [DOI] [Google Scholar]
- Sévin DC, Fuhrer T, Zamboni N, Sauer U, 2016. Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli. Nat. Methods 14. doi: 10.1038/nmeth.4103 [DOI] [PubMed] [Google Scholar]
- Smusz S, Kurczab R, Bojarski AJ, 2013. The influence of the inactives subset generation on the performance of machine learning methods. J. Cheminform 5, 17. doi: 10.1186/1758-2946-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terfloth L, Bienfait B, Gasteiger J, 2007. Ligand-based models for the isoform specificity of cytochrome P450 3A4, 2D6, and 2C9 substrates. J. Chem. Inf. Model 47, 1688–701. doi: 10.1021/ci700010t [DOI] [PubMed] [Google Scholar]
- van der Maaten LJP, Hinton GE, 2008. Visualizing High-Dimensional Data Using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Van Schaftingen E, Rzem R, Marbaix A, Collard F, Veiga-da-Cunha M, Linster CL, 2013. Metabolite proofreading, a neglected aspect of intermediary metabolism. J. Inherit. Metab. Dis 36, 427–34. doi: 10.1007/s10545-012-9571-1 [DOI] [PubMed] [Google Scholar]
- Venkitasubramanian P, Daniels L, Das S, Lamm AS, Rosazza JPN, 2008. Aldehyde oxidoreductase as a biocatalyst: Reductions of vanillic acid. Enzyme Microb. Technol 42, 130–137. doi: 10.1016/j.enzmictec.2007.08.009 [DOI] [PubMed] [Google Scholar]
- Venkitasubramanian P, Daniels L, Rosazza JPN, 2007a. Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme. J. Biol. Chem 282, 478–85. doi: 10.1074/jbc.M607980200 [DOI] [PubMed] [Google Scholar]
- Venkitasubramanian P, Daniels L, Rosazza JPN, 2007b. Biocatalytic reduction of carboxylic acids: mechanism and applications, in: Patel RN (Ed.), Biocatalysis in the Pharmaceutical and Biotechnology Industries. CRC Press, Boca Raton, Florida, pp. 425–440. [Google Scholar]
- Verdel-Aranda K, López-Cortina ST, Hodgson DA, Barona-Gómez F, 2015. Molecular annotation of ketol-acid reductoisomerases from Streptomyces reveals a novel amino acid biosynthesis interlock mediated by enzyme promiscuity. Microb. Biotechnol 8, 239–252. doi: 10.1111/1751-7915.12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wale N, Karypis G, 2009. Target fishing for chemical compounds using target-ligand activity data and ranking based methods. J. Chem. Inf. Model 49, 2190–2201. doi: 10.1021/ci9000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmuth MK, Liao J, Rätsch G, Mathieson M, Putta S, Lemmen C, 2003. Active learning with support vector machines in the drug discovery process. J. Chem. Inf. Comput. Sci 43, 667–673. doi: 10.1021/ci025620t [DOI] [PubMed] [Google Scholar]
- Willett P, 2006. Similarity-based virtual screening using 2D fingerprints. Drug Discov. Today 11, 1046–53. doi: 10.1016/j.drudis.2006.10.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.